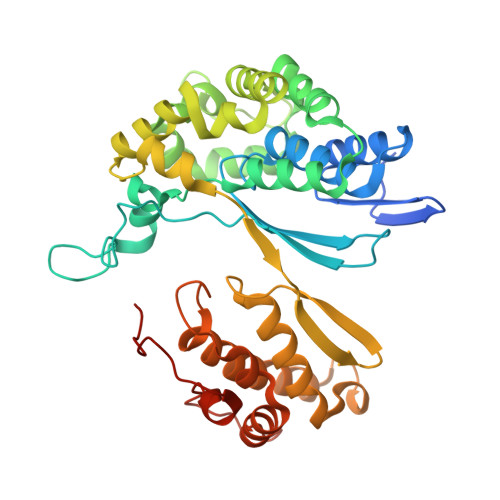
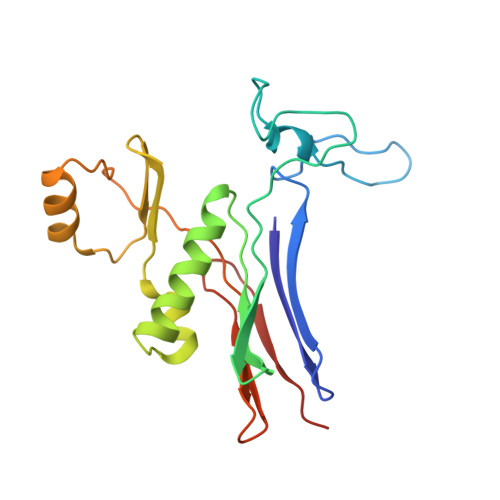
Crystal structure of the halotolerant gamma-glutamyltranspeptidase from Bacillus subtilis in complex with glutamate reveals a unique architecture of the solvent-exposed catalytic pocket
Wada, K., Irie, M., Suzuki, H., Fukuyama, K.(2010) FEBS J 277: 1000-1009
- PubMed: 20088880
- DOI: https://doi.org/10.1111/j.1742-4658.2009.07543.x
- Primary Citation of Related Structures:
3A75 - PubMed Abstract:
gamma-Glutamyltranspeptidase (GGT; EC 2.3.2.2), an enzyme found in organisms from bacteria to mammals and plants, plays a central role in glutathione metabolism. Structural studies of GGTs from Escherichia coli and Helicobacter pylori have revealed detailed molecular mechanisms of catalysis and maturation. In these two GGTs, highly conserved residues form the catalytic pockets, conferring the ability of the loop segment to shield the bound gamma-glutamyl moiety from the solvent. Here, we have examined the Bacillus subtilis GGT, which apparently lacks the amino acids corresponding to the lid-loop that are present in mammalian and plant GGTs as well as in most bacterial GGTs. Another remarkable feature of B. subtilis GGT is its salt tolerance; it retains 86% of its activity even in 3 m NaCl. To better understand these characteristics of B. subtilis GGT, we determined its crystal structure in complex with glutamate, a product of the enzymatic reaction, at 1.95 A resolution. This structure revealed that, unlike the E. coli and H. pylori GGTs, the catalytic pocket of B. subtilis GGT has no segment that covers the bound glutamate; consequently, the glutamate is exposed to solvent. Furthermore, calculation of the electrostatic potential showed that strong acidic patches were distributed on the surface of the B. subtilis GGT, even under high-salt conditions, and this may allow the protein to remain in the hydrated state and avoid self-aggregation. The structural findings presented here have implications for the molecular mechanism of GGT.
- Department of Biological Sciences, Graduate School of Science, Osaka University, Japan.
Organizational Affiliation: