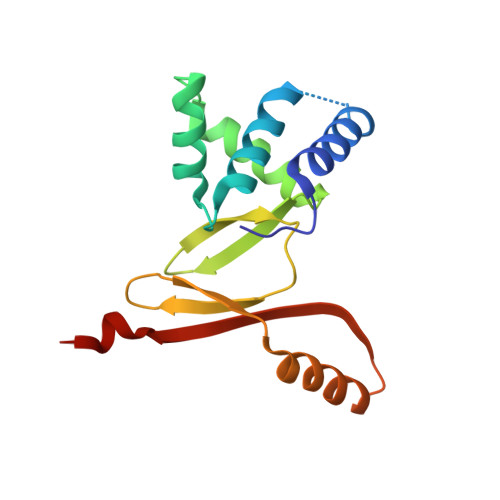
The Structure of the Helicobacter Pylori Ferric Uptake Regulator Fur Reveals Three Functional Metal Binding Sites.
Dian, C., Vitale, S., Leonard, G.A., Bahlawane, C., Fauquant, C., Leduc, D., Muller, C., De Reuse, H., Michaud-Soret, I., Terradot, L.(2011) Mol Microbiol 79: 1260
- PubMed: 21208302
- DOI: https://doi.org/10.1111/j.1365-2958.2010.07517.x
- Primary Citation of Related Structures:
2XIG - PubMed Abstract:
Fur, the ferric uptake regulator, is a transcription factor that controls iron metabolism in bacteria. Binding of ferrous iron to Fur triggers a conformational change that activates the protein for binding to specific DNA sequences named Fur boxes. In Helicobacter pylori, HpFur is involved in acid response and is important for gastric colonization in model animals. Here we present the crystal structure of a functionally active HpFur mutant (HpFur2M; C78S-C150S) bound to zinc. Although its fold is similar to that of other Fur and Fur-like proteins, the crystal structure of HpFur reveals a unique structured N-terminal extension and an unusual C-terminal helix. The structure also shows three metal binding sites: S1 the structural ZnS₄ site previously characterized biochemically in HpFur and the two zinc sites identified in other Fur proteins. Site-directed mutagenesis and spectroscopy analyses of purified wild-type HpFur and various mutants show that the two metal binding sites common to other Fur proteins can be also metallated by cobalt. DNA protection and circular dichroism experiments demonstrate that, while these two sites influence the affinity of HpFur for DNA, only one is absolutely required for DNA binding and could be responsible for the conformational changes of Fur upon metal binding while the other is a secondary site.
- Structural Biology Group, European Synchrotron Radiation Facility, BP 220 F-38043 Grenoble cedex 9, France.
Organizational Affiliation: