Crystallographic Insight Into Collagen Recognition by Discoidin Domain Receptor 2
Carafoli, F., Bihan, D., Stathopoulos, S., Konitsiotis, A.D., Kvansakul, M., Farndale, R.W., Leitinger, B., Hohenester, E.(2009) Structure 17: 1573
- PubMed: 20004161
- DOI: https://doi.org/10.1016/j.str.2009.10.012
- Primary Citation of Related Structures:
2WUH - PubMed Abstract:
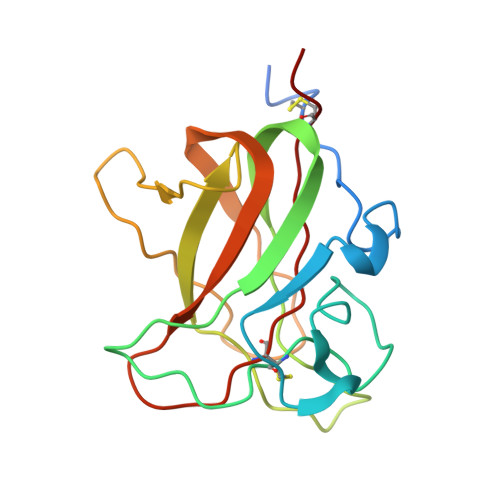
The discoidin domain receptors, DDR1 and DDR2, are widely expressed receptor tyrosine kinases that are activated by triple-helical collagen. They control important aspects of cell behavior and are dysregulated in several human diseases. The major DDR2-binding site in collagens I-III is a GVMGFO motif (O is hydroxyproline) that also binds the matricellular protein SPARC. We have determined the crystal structure of the discoidin domain of human DDR2 bound to a triple-helical collagen peptide. The GVMGFO motifs of two collagen chains are recognized by an amphiphilic pocket delimited by a functionally critical tryptophan residue and a buried salt bridge. Collagen binding results in structural changes of DDR2 surface loops that may be linked to the process of receptor activation. A comparison of the GVMGFO-binding sites of DDR2 and SPARC reveals a striking case of convergent evolution in collagen recognition.
- Department of Life Sciences, Imperial College London, London SW7 2AZ, UK.
Organizational Affiliation: