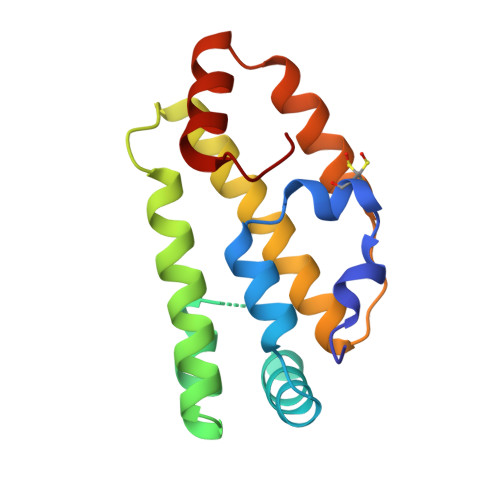
Fatty Acid and Retinoid Binding Proteins Have Distinct Binding Pockets for the Two Types of Cargo
Jordanova, R., Groves, M.R., Kostova, E.B., Woltersdorf, C., Liebau, E., Tucker, P.A.(2009) J Biological Chem 284: 35818
- PubMed: 19828452
- DOI: https://doi.org/10.1074/jbc.M109.022731
- Primary Citation of Related Structures:
2W9Y - PubMed Abstract:
Parasitic nematodes cause serious diseases in humans, animals, and plants. They have limited lipid metabolism and are reliant on lipid-binding proteins to acquire these metabolites from their hosts. Several structurally novel families of lipid-binding proteins in nematodes have been described, including the fatty acid- and retinoid-binding protein family (FAR). In Caenorhabditis elegans, used as a model for studying parasitic nematodes, eight C. elegans FAR proteins have been described. The crystal structure of C. elegans FAR-7 is the first structure of a FAR protein, and it exhibits a novel fold. It differs radically from the mammalian fatty acid-binding proteins and has two ligand binding pockets joined by a surface groove. The first can accommodate the aliphatic chain of fatty acids, whereas the second can accommodate the bulkier retinoids. In addition to demonstrating lipid binding by fluorescence spectroscopy, we present evidence that retinol binding is positively regulated by casein kinase II phosphorylation at a conserved site near the bottom of the second pocket. far-7::GFP (green fluorescent protein) expression shows that it is localized in the head hypodermal syncytia and the excretory cell but that this localization changes under starvation conditions. In conclusion, our study provides the basic structural and functional information for investigation of inhibitors of lipid binding by FAR proteins.
- European Molecular Biology Laboratory, Hamburg Outstation, 22603 Hamburg, Germany.
Organizational Affiliation: