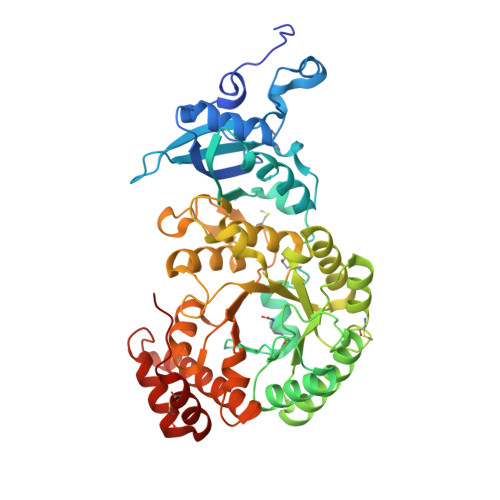
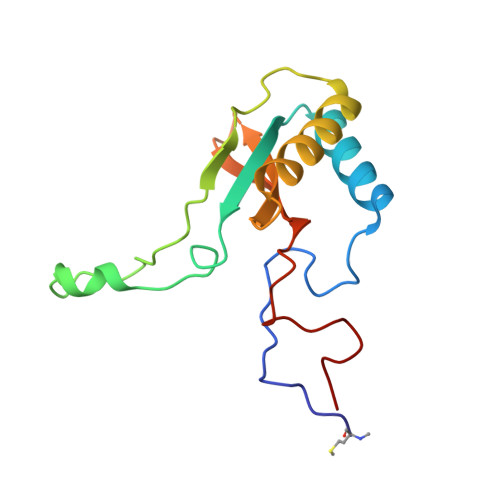
Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2-O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Karkehabadi, S., Satagopan, S., Taylor, T.C., Spreitzer, R.J., Andersson, I.(2007) Biochemistry 46: 11080
- PubMed: 17824672
- DOI: https://doi.org/10.1021/bi701063f
- Primary Citation of Related Structures:
2V63, 2V67, 2V68, 2V69, 2V6A - PubMed Abstract:
The loop between alpha-helix 6 and beta-strand 6 in the alpha/beta-barrel of ribulose-1,5-bisphosphate carboxylase/oxygenase plays a key role in discriminating between CO2 and O2. Genetic screening in Chlamydomonas reinhardtii previously identified a loop-6 V331A substitution that decreases carboxylation and CO2/O2 specificity. Revertant selection identified T342I and G344S substitutions that restore photosynthetic growth by increasing carboxylation and specificity of the V331A enzyme. In numerous X-ray crystal structures, loop 6 is closed or open depending on the activation state of the enzyme and the presence or absence of ligands. The carboxy terminus folds over loop 6 in the closed state. To study the molecular basis for catalysis, directed mutagenesis and chloroplast transformation were used to create T342I and G344S substitutions alone. X-ray crystal structures were then solved for the V331A, V331A/T342I, T342I, and V331A/G344S enzymes, as well as for a D473E enzyme created to assess the role of the carboxy terminus in loop-6 closure. V331A disturbs a hydrophobic pocket, abolishing several van der Waals interactions. These changes are complemented by T342I and G344S, both of which alone cause decreases in CO2/O2 specificity. In the V331A/T342I revertant enzyme, Arg339 main-chain atoms are displaced. In V331A/G344S, alpha-helix 6 is shifted. D473E causes disorder of the carboxy terminus, but loop 6 remains closed. Interactions between a transition-state analogue and several residues are altered in the mutant enzymes. However, active-site Lys334 at the apex of loop 6 has a normal conformation. A variety of subtle interactions must be responsible for catalytic efficiency and CO2/O2 specificity.
- Department of Molecular Biology, Swedish University of Agricultural Sciences, BMC Box 590, 751 24 Uppsala, Sweden.
Organizational Affiliation: