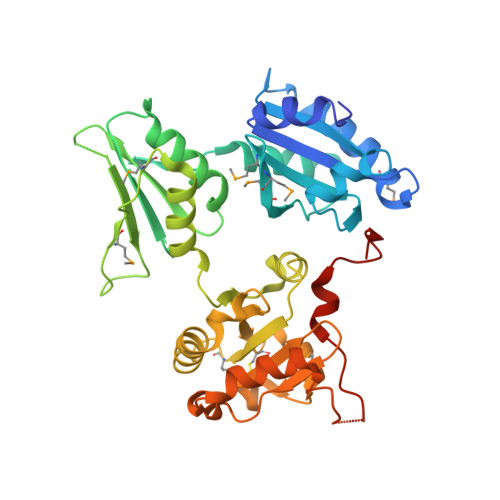
Crystal structure of human ERp44 shows a dynamic functional modulation by its carboxy-terminal tail.
Wang, L.K., Wang, L., Vavassori, S., Li, S.J., Ke, H., Anelli, T., Degano, M., Ronzoni, R., Sitia, R., Sun, F., Wang, C.C.(2008) EMBO Rep
- PubMed: 18552768
- DOI: https://doi.org/10.1038/embor.2008.88
- Primary Citation of Related Structures:
2R2J - PubMed Abstract:
ERp44 mediates thiol-dependent retention in the early secretory pathway, forming mixed disulphides with substrate proteins through its conserved CRFS motif. Here, we present its crystal structure at a resolution of 2.6 A. Three thioredoxin domains-a, b and b'-are arranged in a clover-like structure. A flexible carboxy-terminal tail turns back to the b' and a domains, shielding a hydrophobic pocket in domain b' and a hydrophobic patch around the CRFS motif in domain a. Mutational and functional studies indicate that the C-terminal tail gates the CRFS area and the adjacent hydrophobic pocket, dynamically regulating protein quality control.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: