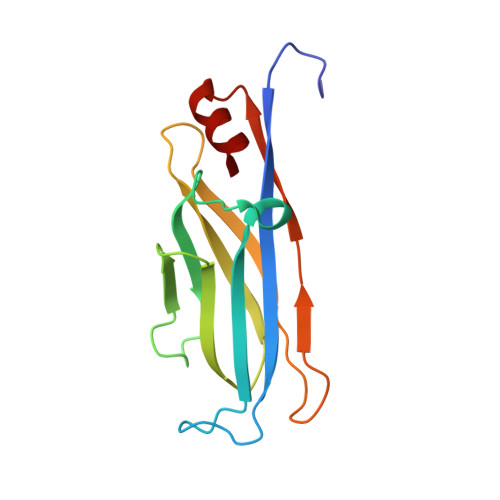
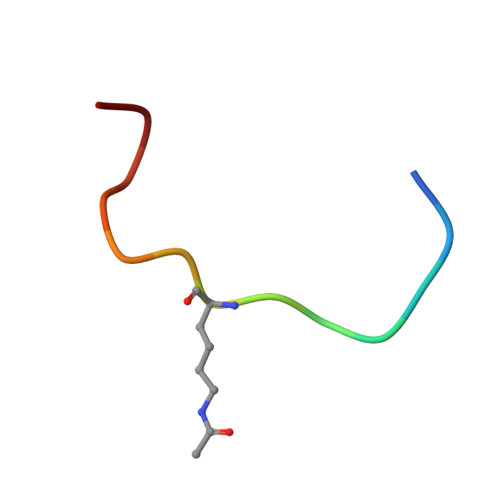
Structural Insights into Histone Crotonyl-Lysine Recognition by the AF9 YEATS Domain.
Zhang, Q., Zeng, L., Zhao, C., Ju, Y., Konuma, T., Zhou, M.M.(2016) Structure 24: 1606-1612
- PubMed: 27545619
- DOI: https://doi.org/10.1016/j.str.2016.05.023
- Primary Citation of Related Structures:
2NDF, 2NDG - PubMed Abstract:
Histone lysine acylations play an important role in the regulation of gene transcription in chromatin. Unlike histone acetyl-lysine, molecular recognition of a recently identified crotonyl-lysine mark is much less understood. Here, we report that the YEATS domain of AF9 preferentially binds crotonyl-lysine over acetyl-lysine in histone H3. Nuclear magnetic resonance structural analysis reveals that crotonyl-lysine of histone H3 lysine 18 is engulfed deep in an aromatic cage of the YEATS domain where the carbonyl oxygen of crotonyl-lysine forms a hydrogen bond with the backbone amide of protein residue Tyr78. The crotonyl-lysine, through its unique electron-rich double-bond side chain, engages π-π aromatic stacking and extended hydrophobic/aromatic interactions with the YEATS domain compared with acetyl-lysine. Our mutational analysis confirmed key protein residues Phe59 and Tyr78 for crotonyl-lysine recognition. Importantly, our findings present a new structural mechanism of protein-protein interactions mediated by histone lysine crotonylation, and show how the cells interpret acyl-lysine marks in different biological contexts.
- The First Hospital and Institute of Epigenetic Medicine, Jilin University, Changchun 130061, China; Department of Structural and Chemical Biology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. Electronic address: qiang.zhang@mssm.edu.
Organizational Affiliation: