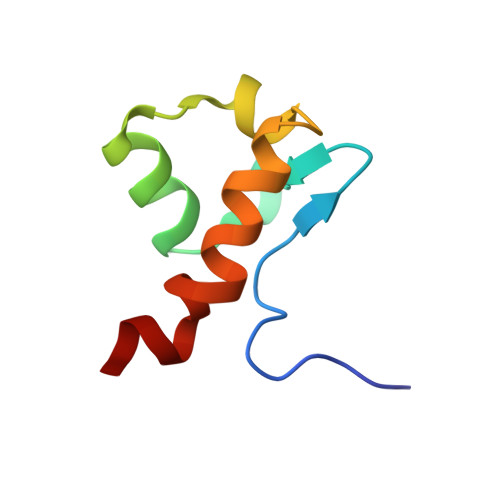
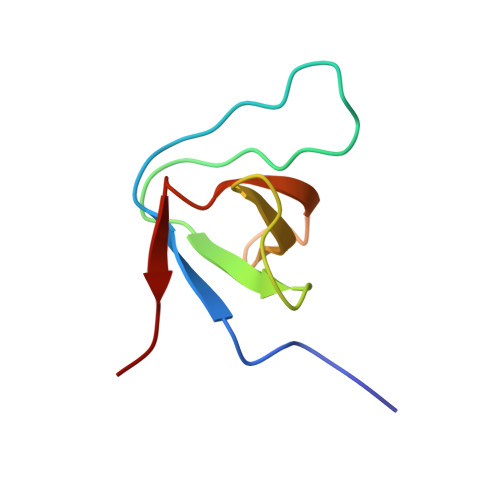
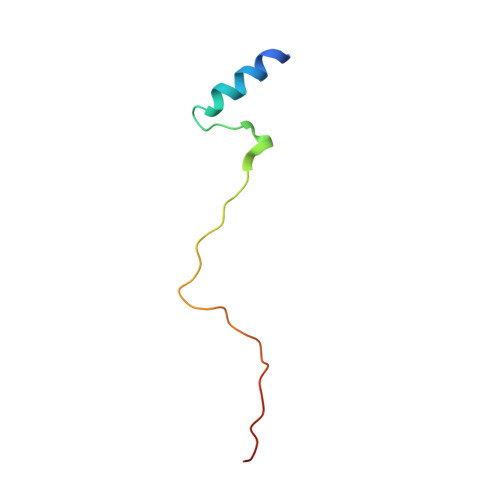
Enterohaemorrhagic Escherichia coli exploits a tryptophan switch to hijack host f-actin assembly.
Aitio, O., Hellman, M., Skehan, B., Kesti, T., Leong, J.M., Saksela, K., Permi, P.(2012) Structure 20: 1692-1703
- PubMed: 22921828
- DOI: https://doi.org/10.1016/j.str.2012.07.015
- Primary Citation of Related Structures:
2LNH - PubMed Abstract:
Intrinsically disordered protein (IDP)-mediated interactions are often characterized by low affinity but high specificity. These traits are essential in signaling and regulation that require reversibility. Enterohaemorrhagic Escherichia coli (EHEC) exploit this situation by commandeering host cytoskeletal signaling to stimulate actin assembly beneath bound bacteria, generating "pedestals" that promote intestinal colonization. EHEC translocates two proteins, EspF(U) and Tir, which form a complex with the host protein IRTKS. The interaction of this complex with N-WASP triggers localized actin polymerization. We show that EspF(U) is an IDP that contains a transiently α-helical N-terminus and dynamic C-terminus. Our structure shows that single EspF(U) repeat forms a high-affinity trimolecular complex with N-WASP and IRTKS. We demonstrate that bacterial and cellular ligands interact with IRTKS SH3 in a similar fashion, but the bacterial protein has evolved to outcompete cellular targets by utilizing a tryptophan switch that offers superior binding affinity enabling EHEC-induced pedestal formation.
- Program in Structural Biology and Biophysics, Institute of Biotechnology, University of Helsinki, FI-00014, Helsinki, Finland.
Organizational Affiliation: