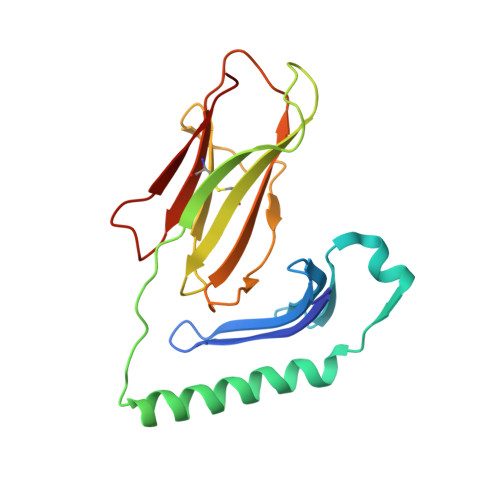
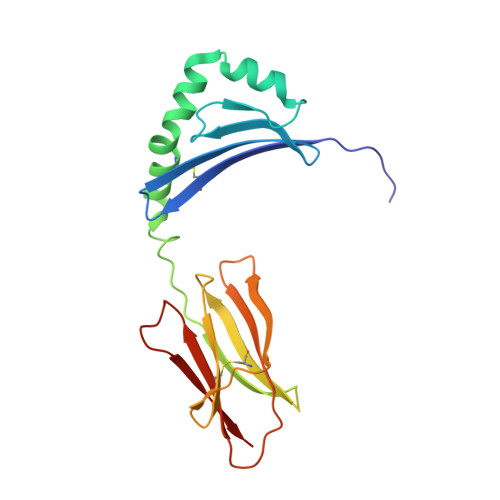
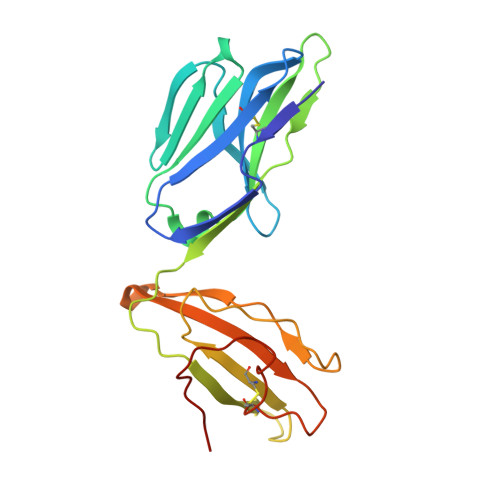
Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor
Deng, L., Langley, R.J., Brown, P.H., Xu, G., Teng, L., Wang, Q., Gonzales, M.I., Callender, G.G., Nishimura, M.I., Topalian, S.L., Mariuzza, R.A.(2007) Nat Immunol 8: 398-408
- PubMed: 17334368
- DOI: https://doi.org/10.1038/ni1447
- Primary Citation of Related Structures:
2IAL, 2IAM, 2IAN - PubMed Abstract:
Structural studies of complexes of T cell receptor (TCR) and peptide-major histocompatibility complex (MHC) have focused on TCRs specific for foreign antigens or native self. An unexplored category of TCRs includes those specific for self determinants bearing alterations resulting from disease, notably cancer. We determined here the structure of a human melanoma-specific TCR (E8) bound to the MHC molecule HLA-DR1 and an epitope from mutant triosephosphate isomerase. The structure had features intermediate between 'anti-foreign' and autoimmune TCR-peptide-MHC class II complexes that may reflect the hybrid nature of altered self. E8 manifested very low affinity for mutant triosephosphate isomerase-HLA-DR1 despite the highly tumor-reactive properties of E8 cells. A second TCR (G4) had even lower affinity but underwent peptide-specific formation of dimers, suggesting this as a mechanism for enhancing low-affinity TCR-peptide-MHC interactions for T cell activation.
- Center for Advanced Research in Biotechnology, WM Keck Laboratory for Structural Biology, University of Maryland Biotechnology Institute, Rockville, Maryland 20850, USA.
Organizational Affiliation: