Structure of dNTP-inducible dNTP triphosphohydrolase: insight into broad specificity for dNTPs and triphosphohydrolase-type hydrolysis
Kondo, N., Nakagawa, N., Ebihara, A., Chen, L., Liu, Z.-J., Wang, B.-C., Yokoyama, S., Kuramitsu, S., Masui, R.(2007) Acta Crystallogr D Biol Crystallogr 63: 230-239
- PubMed: 17242516
- DOI: https://doi.org/10.1107/S0907444906049262
- Primary Citation of Related Structures:
2DQB - PubMed Abstract:
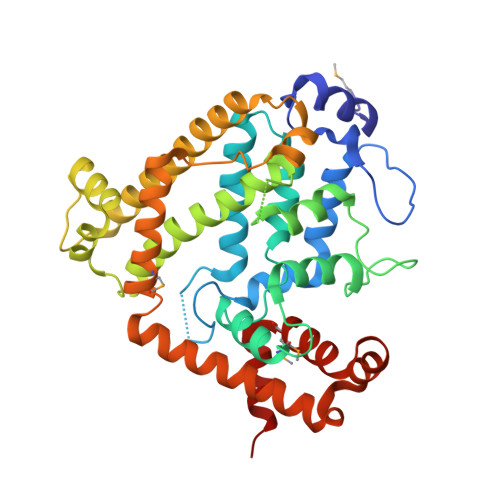
Deoxyribonucleoside triphosphate triphosphohydrolase from Thermus thermophilus (Tt-dNTPase) has a unique regulatory mechanism for the degradation of deoxyribonucleoside triphosphates (dNTPs). Whereas the Escherichia coli homologue specifically hydrolyzes dGTP alone, dNTPs act as both substrate and activator for Tt-dNTPase. Here, the crystal structure of Tt-dNTPase has been determined at 2.2 A resolution, representing the first report of the tertiary structure of a dNTPase homologue belonging to the HD superfamily, a diverse group of metal-dependent phosphohydrolases that includes a variety of uncharacterized proteins. This enzyme forms a homohexamer as a double ring of trimers. The subunit is composed of 19 alpha-helices; the inner six helices include the region annotated as the catalytic domain of the HD superfamily. Structural comparison with other HD-superfamily proteins indicates that a pocket at the centre of the inner six helices, formed from highly conserved charged residues clustered around a bound magnesium ion, constitutes the catalytic site. Tt-dNTPase also hydrolyzed noncanonical dNTPs, but hardly hydrolyzed dNDP and dNMP. The broad substrate specificity for different dNTPs might be rationalized by the involvement of a flexible loop during molecular recognition of the base moiety. Recognition of the triphosphate moiety crucial for the activity might be attained by highly conserved positively charged residues. The possible mode of dNTP binding is discussed in light of the structure.
- Department of Biological Sciences, Graduate School of Science, Osaka University, 1-1 Machikaneyama-cho, Toyonaka, Osaka 560-0043, Japan.
Organizational Affiliation: