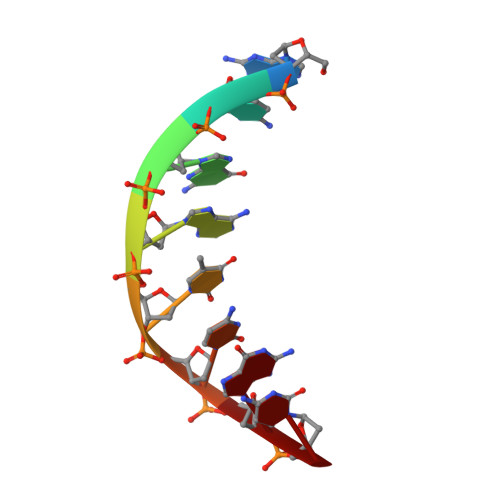
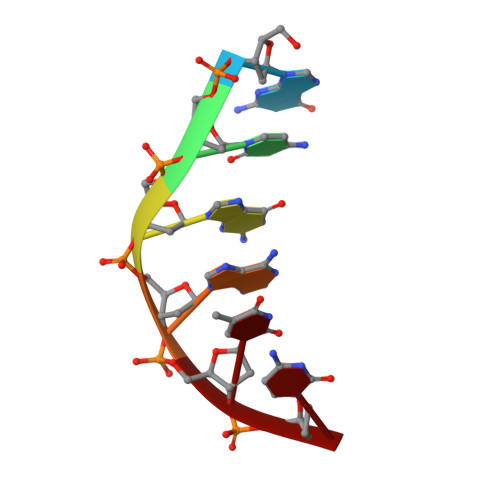
DNase I-induced DNA conformation. 2 A structure of a DNase I-octamer complex
Lahm, A., Suck, D.(1991) J Mol Biology 222: 645-667
- PubMed: 1748997
- DOI: https://doi.org/10.1016/0022-2836(91)90502-w
- Primary Citation of Related Structures:
2DNJ - PubMed Abstract:
The structure of a complex between DNase I and d(GCGATCGC)2 has been solved by molecular replacement and refined to an R-factor of 0.174 for all data between 6 and 2 A resolution. The nicked octamer duplexes have lost a dinucleotide from the 3' ends of one strand and are hydrogen-bonded across a 2-fold axis to form a quasi-continuous double helix of 14 base-pairs. DNase I is bound in the minor groove of the B-type DNA duplex forming contacts in and along both sides of the minor groove extending over a total of six base-pairs. As a consequence of binding of DNase I to the DNA-substrate the minor groove opens by about 3 A and the duplex bends towards the major groove by about 20 degrees. Apart from these more global distortions the bound duplex also shows significant deviations in local geometry. A major cause for the observed perturbations in the DNA conformation seems to be the stacking type interaction of a tyrosine ring (Y76) with a deoxyribose. In contrast, the enzyme structure is nearly unchanged compared to free DNase I (0.49 A root-mean-square deviations for main-chain atoms) thus providing a rigid framework to which the DNA substrate has to adapt on binding. These results confirm the hypothesis that groove width and stiffness are major factors determining the global sequence dependence of the enzyme's cutting rates. The nicked octamer present in the crystals did not allow us to draw detailed conclusions about the catalytic mechanism but confirmed the location of the active site near H134 on top of the central beta-sheets. A second cut of the DNA induced by diffusion of Mn2+ into the crystals may suggest the presence of a secondary active site in DNase I.
- European Molecular Biology Laboratory, Biological Structures Division, Heidelberg, Germany.
Organizational Affiliation: