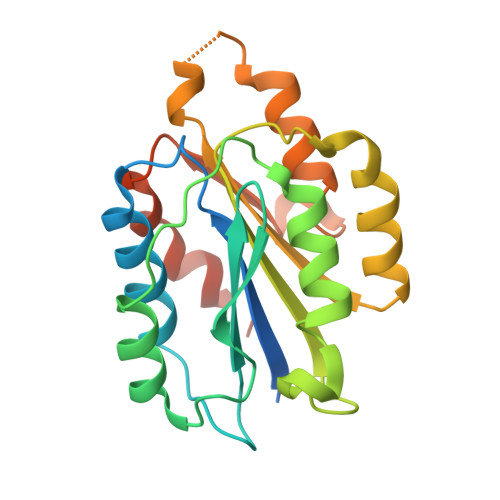
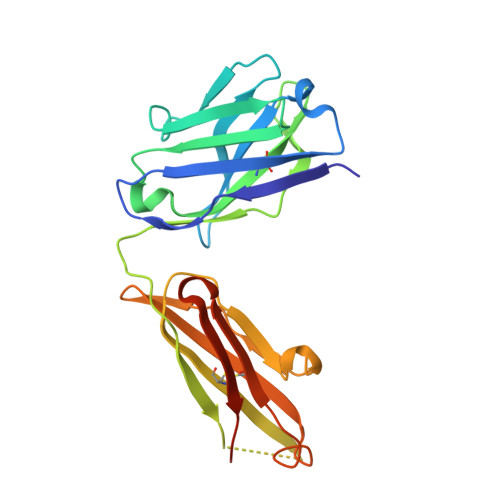
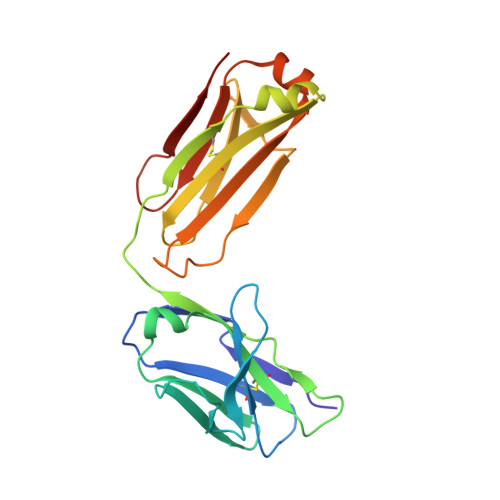
Affinity enhancement of an in vivo matured therapeutic antibody using structure-based computational design
Clark, L.A., Boriack-Sjodin, P.A., Eldredge, J., Fitch, C., Friedman, B., Hanf, K.J., Jarpe, M., Liparoto, S.F., Li, Y., Lugovskoy, A., Miller, S., Rushe, M., Sherman, W., Simon, K., Van Vlijmen, H.(2006) Protein Sci 15: 949-960
- PubMed: 16597831
- DOI: https://doi.org/10.1110/ps.052030506
- Primary Citation of Related Structures:
2B2X - PubMed Abstract:
Improving the affinity of a high-affinity protein-protein interaction is a challenging problem that has practical applications in the development of therapeutic biomolecules. We used a combination of structure-based computational methods to optimize the binding affinity of an antibody fragment to the I-domain of the integrin VLA1. Despite the already high affinity of the antibody (Kd approximately 7 nM) and the moderate resolution (2.8 A) of the starting crystal structure, the affinity was increased by an order of magnitude primarily through a decrease in the dissociation rate. We determined the crystal structure of a high-affinity quadruple mutant complex at 2.2 A. The structure shows that the design makes the predicted contacts. Structural evidence and mutagenesis experiments that probe a hydrogen bond network illustrate the importance of satisfying hydrogen bonding requirements while seeking higher-affinity mutations. The large and diverse set of interface mutations allowed refinement of the mutant binding affinity prediction protocol and improvement of the single-mutant success rate. Our results indicate that structure-based computational design can be successfully applied to further improve the binding of high-affinity antibodies.
- Biogen Idec, Inc., Cambridge, Massachusetts 02142, USA. louie@alumni.northwestern.edu
Organizational Affiliation: