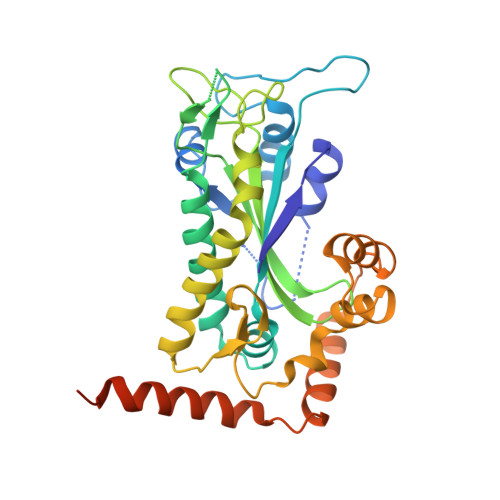
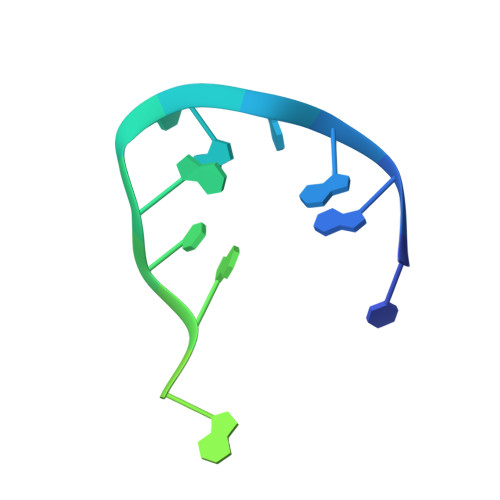
Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the f factor relaxase.
Larkin, C., Datta, S., Harley, M.J., Anderson, B.J., Ebie, A., Hargreaves, V., Schildbach, J.F.(2005) Structure 13: 1533-1544
- PubMed: 16216584
- DOI: https://doi.org/10.1016/j.str.2005.06.013
- Primary Citation of Related Structures:
2A0I - PubMed Abstract:
The TraI protein of conjugative plasmid F factor binds and cleaves a single-stranded region of the plasmid prior to transfer to a recipient. TraI36, an N-terminal TraI fragment, binds ssDNA with a subnanomolar K(D) and remarkable sequence specificity. The structure of the TraI36 Y16F variant bound to ssDNA reveals specificity determinants, including a ssDNA intramolecular 3 base interaction and two pockets within the protein's binding cleft that accommodate bases in a knob-into-hole fashion. Mutagenesis results underscore the intricate design of the binding site, with the greatest effects resulting from substitutions for residues that both contact ssDNA and stabilize protein structure. The active site architecture suggests that the bound divalent cation, which is essential for catalysis, both positions the DNA by liganding two oxygens of the scissile phosphate and increases the partial positive charge on the phosphorus to enhance nucleophilic attack.
- Department of Biology, The Johns Hopkins University, 3400 North Charles Street, Baltimore, Maryland 21218, USA.
Organizational Affiliation: