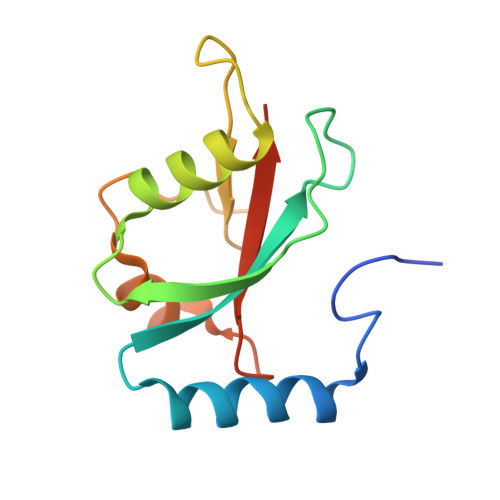
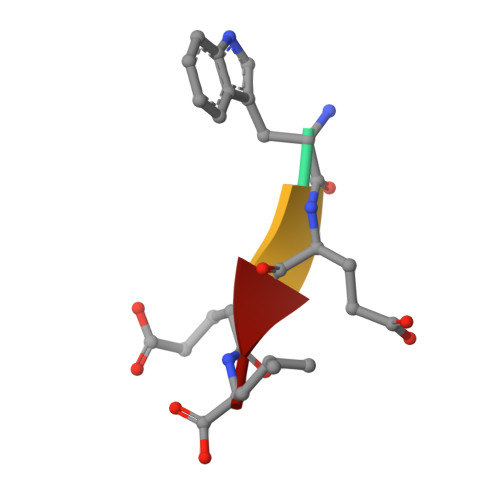
Structural basis of target recognition by Atg8/LC3 during selective autophagy
Noda, N.N., Kumeta, H., Nakatogawa, H., Satoo, K., Adachi, W., Ishii, J., Fujioka, Y., Ohsumi, Y., Inagaki, F.(2008) Genes Cells 13: 1211-1218
- PubMed: 19021777
- DOI: https://doi.org/10.1111/j.1365-2443.2008.01238.x
- Primary Citation of Related Structures:
2ZPN - PubMed Abstract:
Autophagy is a non-selective bulk degradation process in which isolation membranes enclose a portion of cytoplasm to form double-membrane vesicles, called autophagosomes, and deliver their inner constituents to the lytic compartments. Recent studies have also shed light on another mode of autophagy that selectively degrades various targets. Yeast Atg8 and its mammalian homologue LC3 are ubiquitin-like modifiers that are localized on isolation membranes and play crucial roles in the formation of autophagosomes. These proteins are also involved in selective incorporation of specific cargo molecules into autophagosomes, in which Atg8 and LC3 interact with Atg19 and p62, receptor proteins for vacuolar enzymes and disease-related protein aggregates, respectively. Using X-ray crystallography and NMR, we herein report the structural basis for Atg8-Atg19 and LC3-p62 interactions. Remarkably, Atg8 and LC3 were shown to interact with Atg19 and p62, respectively, in a quite similar manner: they recognized the side-chains of Trp and Leu in a four-amino acid motif, WXXL, in Atg19 and p62 using hydrophobic pockets conserved among Atg8 homologues. Together with mutational analyses, our results show the fundamental mechanism that allows Atg8 homologues, in association with WXXL-containing proteins, to capture specific cargo molecules, thereby endowing isolation membranes and/or their assembly machineries with target selectivity.
- Department of Structural Biology, Graduate School of Pharmaceutical Sciences, Hokkaido University, Kita-ku, Sapporo, Japan.
Organizational Affiliation: