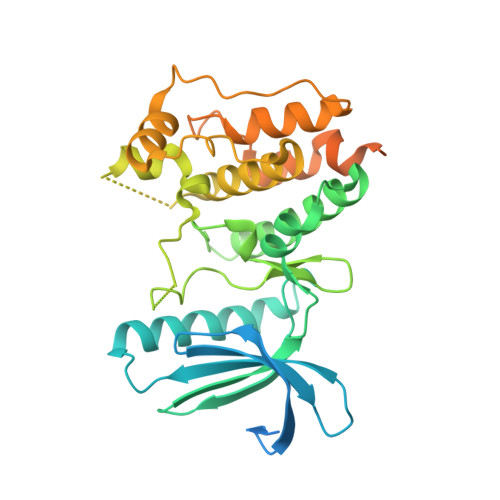
Crystal structure of the catalytic domain of the mitotic checkpoint kinase Mps1 in complex with SP600125.
Chu, M.L., Chavas, L.M., Douglas, K.T., Eyers, P.A., Tabernero, L.(2008) J Biological Chem 283: 21495-21500
- PubMed: 18480048
- DOI: https://doi.org/10.1074/jbc.M803026200
- Primary Citation of Related Structures:
2ZMC, 2ZMD - PubMed Abstract:
Chromosomal instability can result from defective control of checkpoints and is associated with malignant cell growth. Monopolar spindle 1 (Mps1) is a dual-specificity protein kinase that has important roles in the prevention of aneuploidy during the cell cycle and might therefore be a potential target for new therapeutic agents in the treatment of cancer. To gain insights into the molecular mechanism of Mps1 inhibition by small molecules, we determined the x-ray structure of Mps1, both alone and in complex with the ATP-competitive inhibitor SP600125. Mps1 adopts a classic protein kinase fold, with the inhibitor sitting in the ATP-binding site where it is stabilized by hydrophobic interactions. We identified a secondary pocket, not utilized by SP600125, which might be exploited for the rational design of specific Mps1 inhibitors. These structures provide important insights into the interaction of this protein kinase with small molecules and suggest potential mechanisms for Mps1 regulation.
- School of Pharmacy and Pharmaceutical Sciences, Stopford Building, University of Manchester, Manchester, UK.
Organizational Affiliation: