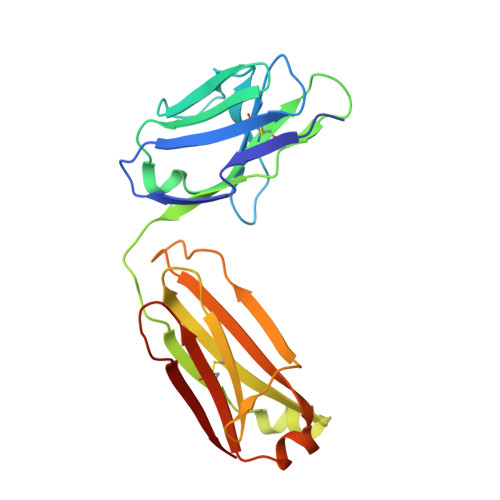
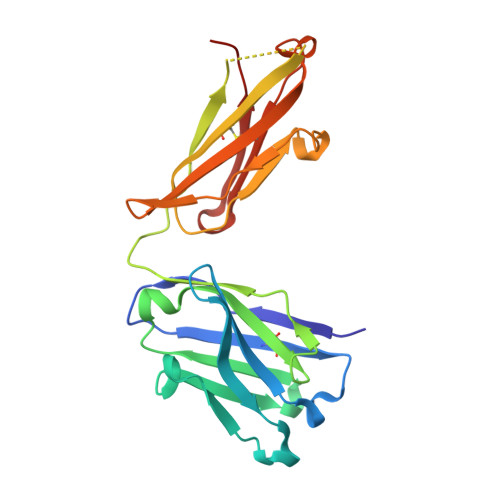
An insight into the thermodynamic characteristics of human thrombopoietin complexation with TN1 antibody.
Arai, S., Shibazaki, C., Adachi, M., Honjo, E., Tamada, T., Maeda, Y., Tahara, T., Kato, T., Miyazaki, H., Blaber, M., Kuroki, R.(2016) Protein Sci 25: 1786-1796
- PubMed: 27419667
- DOI: https://doi.org/10.1002/pro.2985
- Primary Citation of Related Structures:
2ZKH - PubMed Abstract:
Human thrombopoietin (hTPO) primarily stimulates megakaryocytopoiesis and platelet production and is neutralized by the mouse TN1 antibody. The thermodynamic characteristics of TN1 antibody-hTPO complexation were analyzed by isothermal titration calorimetry (ITC) using an antigen-binding fragment (Fab) derived from the TN1 antibody (TN1-Fab). To clarify the mechanism by which hTPO is recognized by TN1-Fab the conformation of free TN1-Fab was determined to a resolution of 2.0 Å using X-ray crystallography and compared with the hTPO-bound form of TN1-Fab determined by a previous study. This structural comparison revealed that the conformation of TN1-Fab does not substantially change after hTPO binding and a set of 15 water molecules is released from the antigen-binding site (paratope) of TN1-Fab upon hTPO complexation. Interestingly, the heat capacity change (ΔCp) measured by ITC (-1.52 ± 0.05 kJ mol(-1) K(-1) ) differed significantly from calculations based upon the X-ray structure data of the hTPO-bound and unbound forms of TN1-Fab (-1.02 ∼ 0.25 kJ mol(-1) K(-1) ) suggesting that hTPO undergoes an induced-fit conformational change combined with significant desolvation upon TN1-Fab binding. The results shed light on the structural biology associated with neutralizing antibody recognition.
- Quantum Beam Science Research Directorate, National Institutes for Quantum and Radiological Science and Technology, 2-4 Shirakata, Tokai, Ibaraki, 319-1106, Japan. arai.shigeki@qst.go.jp.
Organizational Affiliation: