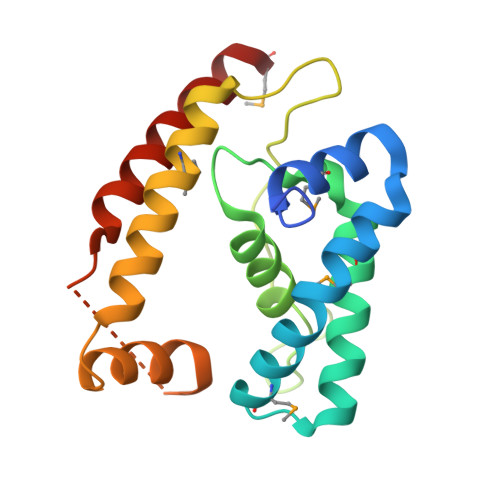
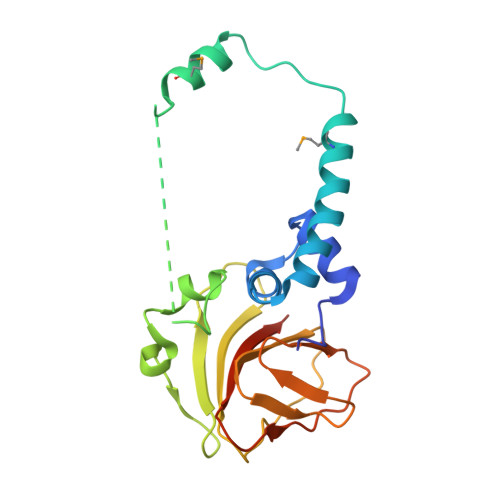
A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria.
Campbell, E.A., Greenwell, R., Anthony, J.R., Wang, S., Lim, L., Das, K., Sofia, H.J., Donohue, T.J., Darst, S.A.(2007) Mol Cell 27: 793-805
- PubMed: 17803943
- DOI: https://doi.org/10.1016/j.molcel.2007.07.009
- Primary Citation of Related Structures:
2Q1Z, 2Z2S - PubMed Abstract:
A transcriptional response to singlet oxygen in Rhodobacter sphaeroides is controlled by the group IV sigma factor sigma(E) and its cognate anti-sigma ChrR. Crystal structures of the sigma(E)/ChrR complex reveal a modular, two-domain architecture for ChrR. The ChrR N-terminal anti-sigma domain (ASD) binds a Zn(2+) ion, contacts sigma(E), and is sufficient to inhibit sigma(E)-dependent transcription. The ChrR C-terminal domain adopts a cupin fold, can coordinate an additional Zn(2+), and is required for the transcriptional response to singlet oxygen. Structure-based sequence analyses predict that the ASD defines a common structural fold among predicted group IV anti-sigmas. These ASDs are fused to diverse C-terminal domains that are likely involved in responding to specific environmental signals that control the activity of their cognate sigma factor.
- The Rockefeller University, 1230 York Avenue, New York, NY 10021, USA.
Organizational Affiliation: