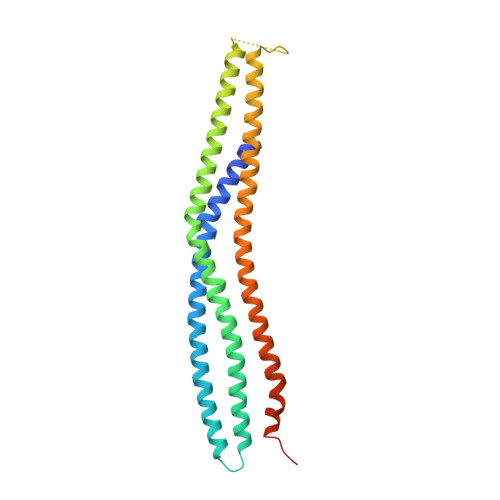
Structural Basis for Complex Formation between Human Irsp53 and the Translocated Intimin Receptor Tir of Enterohemorrhagic E. Coli.
De Groot, J.C., Schluter, K., Carius, Y., Quedenau, C., Vingadassalom, D., Faix, J., Weiss, S.M., Reichelt, J., Standfuss-Gabisch, C., Lesser, C.F., Leong, J.M., Heinz, D.W., Bussow, K., Stradal, T.E.B.(2011) Structure 19: 1294
- PubMed: 21893288
- DOI: https://doi.org/10.1016/j.str.2011.06.015
- Primary Citation of Related Structures:
2YKT - PubMed Abstract:
Actin assembly beneath enterohemorrhagic E. coli (EHEC) attached to its host cell is triggered by the intracellular interaction of its translocated effector proteins Tir and EspF(U) with human IRSp53 family proteins and N-WASP. Here, we report the structure of the N-terminal I-BAR domain of IRSp53 in complex with a Tir-derived peptide, in which the homodimeric I-BAR domain binds two Tir molecules aligned in parallel. This arrangement provides a protein scaffold linking the bacterium to the host cell's actin polymerization machinery. The structure uncovers a specific peptide-binding site on the I-BAR surface, conserved between IRSp53 and IRTKS. The Tir Asn-Pro-Tyr (NPY) motif, essential for pedestal formation, is specifically recognized by this binding site. The site was confirmed by mutagenesis and in vivo-binding assays. It is possible that IRSp53 utilizes the NPY-binding site for additional interactions with as yet unknown partners within the host cell.
- Division of Structural Biology, Helmholtz Centre for Infection Research, 38124 Braunschweig, Germany.
Organizational Affiliation: