Structural Characterization and Membrane Localization of Exsb from the Type III Secretion System (T3Ss) of Pseudomonas Aeruginosa
Izore, T., Perdu, C., Job, V., Atree, I., Faudry, E., Dessen, A.(2011) J Mol Biology 413: 236
- PubMed: 21839744
- DOI: https://doi.org/10.1016/j.jmb.2011.07.043
- Primary Citation of Related Structures:
2YJL - PubMed Abstract:
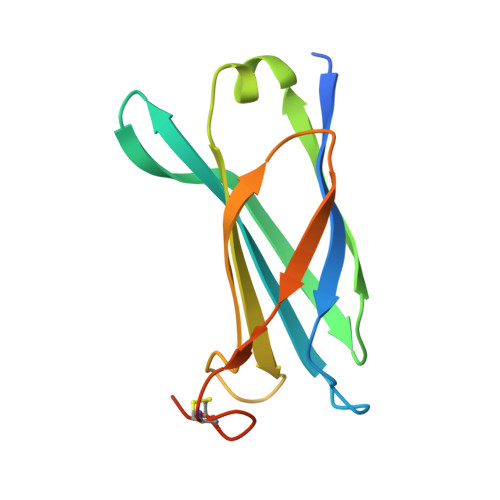
Pseudomonas aeruginosa is an opportunistic human pathogen that employs a finely tuned type III secretion system (T3SS) to inject toxins directly into the cytoplasm of target cells. ExsB is a 15.6-kDa protein encoded in a T3SS transcription regulation operon that displays high sequence similarity to YscW, a lipoprotein from Yersinia spp. whose genetic neighborhood also involves a transcriptional regulator, and has been shown to play a role in the stabilization of the outer membrane ring of the T3SS. Here, we show that ExsB is expressed in P. aeruginosa upon induction of the T3SS, and subcellular fractionation studies reveal that it is associated with the outer membrane. The high-resolution crystal structure of ExsB shows that it displays a compact β-sandwich fold with interdependent β-sheets. ExsB possesses a large patch of basic residues that could play a role in membrane recognition, and its structure is distinct from that of MxiM, a lipoprotein involved in secretin stabilization in Shigella, as well as from those of Pil lipoproteins involved in pilus biogenesis. These results reveal that small lipoproteins involved in formation of the outer membrane secretin ring display clear structural differences that may be related to the different functions they play in these systems.
- Bacterial Pathogenesis Group, Institut de Biologie Structurale (IBS), Université Grenoble I, France.
Organizational Affiliation: