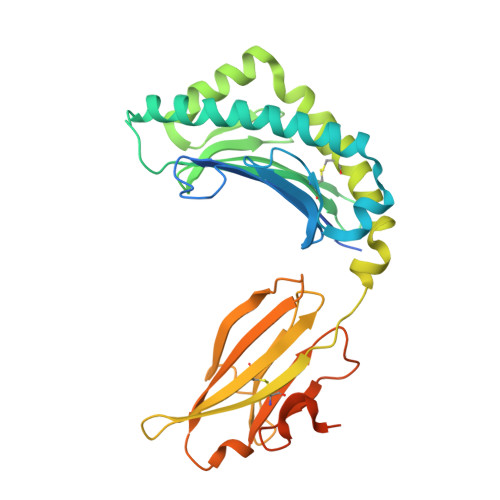
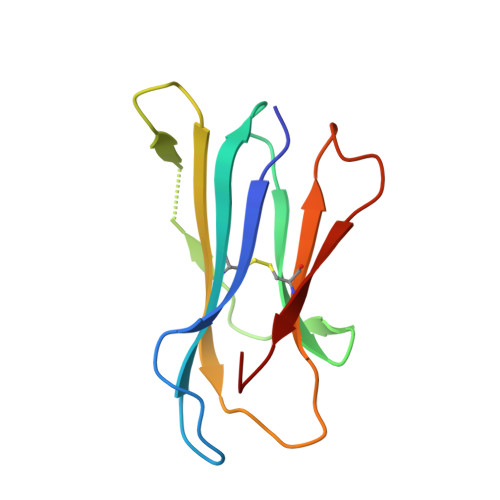
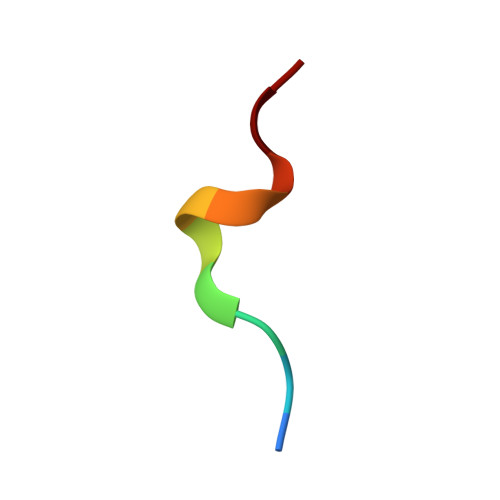Expression levels of MHC class I molecules are inversely correlated with promiscuity of peptide binding.
Chappell, P., Meziane, e.l..K., Harrison, M., Magiera, L., Hermann, C., Mears, L., Wrobel, A.G., Durant, C., Nielsen, L.L., Buus, S., Ternette, N., Mwangi, W., Butter, C., Nair, V., Ahyee, T., Duggleby, R., Madrigal, A., Roversi, P., Lea, S.M., Kaufman, J.(2015) Elife 4: e05345-e05345
- PubMed: 25860507
- DOI: https://doi.org/10.7554/eLife.05345
- Primary Citation of Related Structures:
2YEZ, 4CVX, 4CVZ, 4CW1, 4D0B, 4D0C, 4D0D - PubMed Abstract:
Highly polymorphic major histocompatibility complex (MHC) molecules are at the heart of adaptive immune responses, playing crucial roles in many kinds of disease and in vaccination. We report that breadth of peptide presentation and level of cell surface expression of class I molecules are inversely correlated in both chickens and humans. This relationship correlates with protective responses against infectious pathogens including Marek's disease virus leading to lethal tumours in chickens and human immunodeficiency virus infection progressing to AIDS in humans. We propose that differences in peptide binding repertoire define two groups of MHC class I molecules strategically evolved as generalists and specialists for different modes of pathogen resistance. We suggest that differences in cell surface expression level ensure the development of optimal peripheral T cell responses. The inverse relationship of peptide repertoire and expression is evidently a fundamental property of MHC molecules, with ramifications extending beyond immunology and medicine to evolutionary biology and conservation.
- Sir William Dunn School of Pathology, University of Oxford, Oxford, United Kingdom.
Organizational Affiliation: