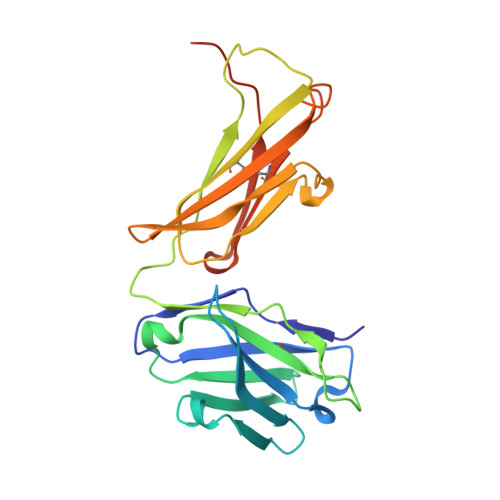
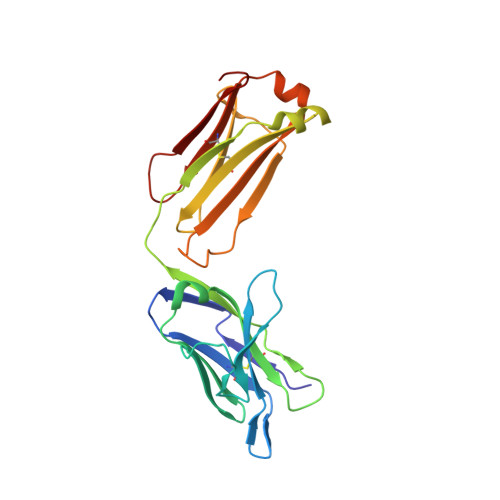
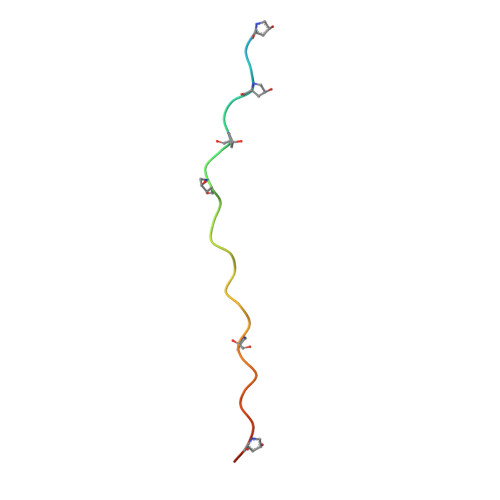
Crystal Structure of an Arthritogenic Anticollagen Immune Complex.
Dobritzsch, D., Lindh, I., Uysal, H., Nandakumar, K.S., Burkhardt, H., Schneider, G., Holmdahl, R.(2011) Arthritis Rheum 63: 3740
- PubMed: 22127694
- DOI: https://doi.org/10.1002/art.30611
- Primary Citation of Related Structures:
2Y5T - PubMed Abstract:
In rheumatoid arthritis, joint inflammation and cartilage destruction are mediated by autoantibodies directed to various self antigens. Type II collagen (CII)-specific antibodies are likely to play a role in this process and have been shown to induce experimental arthritis in susceptible animals. The purpose of this study was to reveal how arthritogenic autoantibodies recognize native CII in its triple-helical conformation. Site-directed mutagenesis and crystallographic studies were performed to reveal crucial contact points between the CII antibody and the triple-helical CII peptide. The crystal structure of a pathogenic autoantibody bound to a major triple-helical epitope present on CII was determined, allowing a first and detailed description of the interactions within an arthritogenic complex that is frequently occurring in both mice and humans with autoimmune arthritis. The crystal structure emphasizes the role of arginine residues located in a commonly recognized motif on CII and reveals that germline-encoded elements are involved in the interaction with the epitope. The crystal structure of an arthritogenic antibody binding a triple-helical epitope on CII indicates a crucial role of germline-encoded and arginine residues as the target structures.
- Karolinska Institute, Stockholm, Sweden.
Organizational Affiliation: