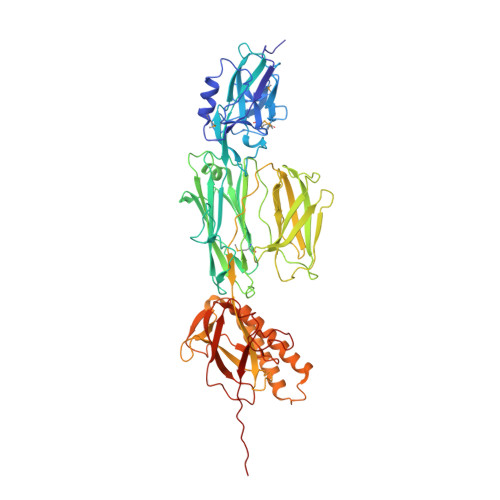
The Full-Length Streptococcus Pneumoniae Major Pilin Rrgb Crystallizes in a Fibre-Like Structure, which Presents the D1 Isopeptide Bond and Provides Details on the Mechanism of Pilus Polymerization.
El Mortaji, L., Contreras-Martel, C., Moschioni, M., Ferlenghi, I., Manzano, C., Vernet, T., Dessen, A., Di Guilmi, A.M.(2012) Biochem J 441: 833
- PubMed: 22013894
- DOI: https://doi.org/10.1042/BJ20111397
- Primary Citation of Related Structures:
2Y1V - PubMed Abstract:
RrgB is the major pilin which forms the pneumococcal pilus backbone. We report the high-resolution crystal structure of the full-length form of RrgB containing the IPQTG sorting motif. The RrgB fold is organized into four distinct domains, D1-D4, each of which is stabilized by an isopeptide bond. Crystal packing revealed a head-to-tail organization involving the interaction of the IPQTG motif into the D1 domain of two successive RrgB monomers. This fibrillar assembly, which fits into the electron microscopy density map of the native pilus, probably induces the formation of the D1 isopeptide bond as observed for the first time in the present study, since neither in published structures nor in soluble RrgB produced in Escherichia coli or in Streptococcus pneumoniae is the D1 bond present. Experiments performed in live bacteria confirmed that the intermolecular bond linking the RrgB subunits takes place between the IPQTG motif of one RrgB subunit and the Lys183 pilin motif residue of an adjacent RrgB subunit. In addition, we present data indicating that the D1 isopeptide bond is involved in RrgB stabilization. In conclusion, the crystal RrgB fibre is a compelling model for deciphering the molecular details required to generate the pneumococcal pilus.
- CEA, Institut de Biologie Structurale Jean-Pierre Ebel, F-38027 Grenoble, France.
Organizational Affiliation: