Structural Elucidation of the Mechanistic Basis of Degeneracy in the Primary Humoral Response.
Khan, T., Salunke, D.M.(2012) J Immunol 188: 1819
- PubMed: 22266283
- DOI: https://doi.org/10.4049/jimmunol.1102701
- Primary Citation of Related Structures:
2XZQ, 2Y06, 2Y07, 2Y36, 4A6Y - PubMed Abstract:
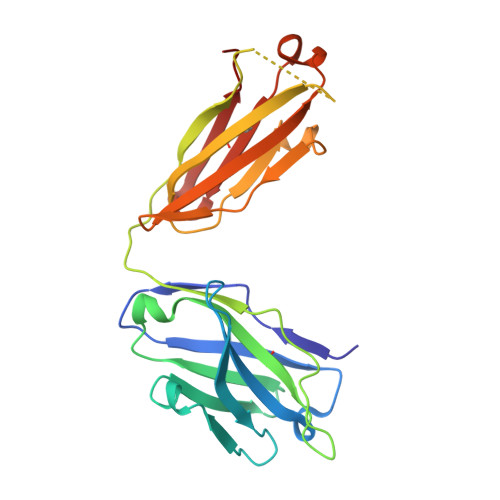
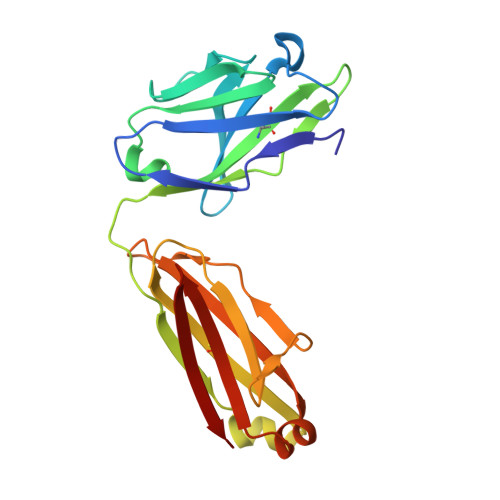
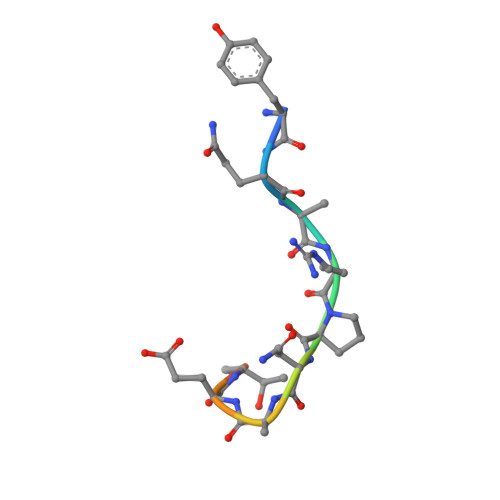
The mechanistic basis for efficient combating of the infinite range of foreign Ags by the limited repertoire of naive Abs expressed on primary B cell surfaces during their first encounter was addressed through elegantly designed crystallographic analyses. Resolution of the discrepancy arising from the limited number of possible germline Ab receptors on primary B cells for recognizing the unlimited pool of possible Ags has been attempted by invoking the degenerate recognition potential of the germline Abs. Structural analyses of germline mAb BBE6.12H3 in an Ag-free state, as well as bound to four different peptide Ags, established the correlation of its degenerate specificity with conformational versatility of the paratope. Six distinct paratope topologies observed for a single germline mAb provided a quantitative description of the primary Ag recognition repertoire at the tertiary structural level. Each of the four different peptide Ags was bound specifically to a distinct conformation of the paratope, which was also different from that of the Ag-free states of the same germline mAb. A minimal conserved motif in the pristine Ag-combining site essential for multispecificity and Ag binding-mediated change in the elbow angle of Fab was also discernible. It is proposed that the generation of a primary Ab repertoire involves large, yet finite, germline Ab clones, each capable of adopting discrete conformations, which in turn exhibit diverse binding modes.
- Structural Biology Unit, National Institute of Immunology, New Delhi 110 067, India.
Organizational Affiliation: