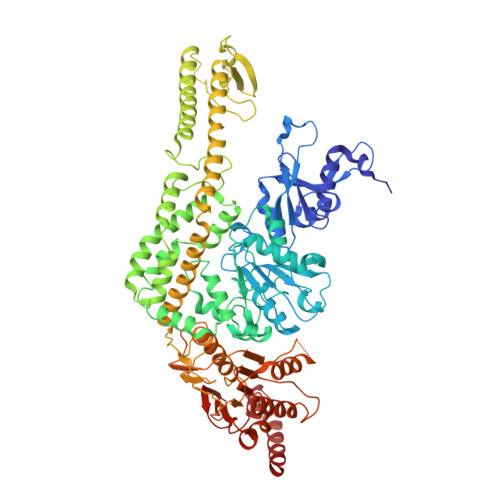
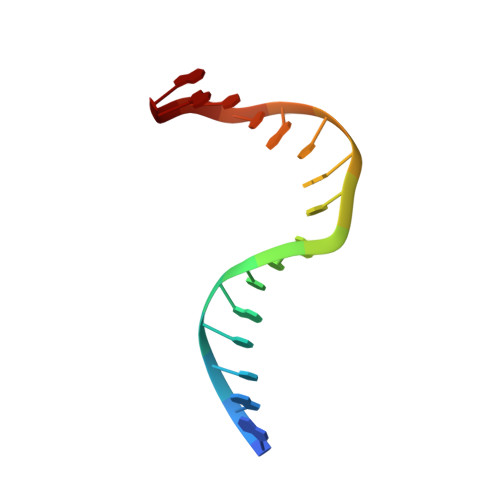
Magnesium coordination controls the molecular switch function of DNA mismatch repair protein MutS.
Lebbink, J.H., Fish, A., Reumer, A., Natrajan, G., Winterwerp, H.H., Sixma, T.K.(2010) J Biological Chem 285: 13131-13141
- PubMed: 20167596
- DOI: https://doi.org/10.1074/jbc.M109.066001
- Primary Citation of Related Structures:
2WTU, 3K0S - PubMed Abstract:
The DNA mismatch repair protein MutS acts as a molecular switch. It toggles between ADP and ATP states and is regulated by mismatched DNA. This is analogous to G-protein switches and the regulation of their "on" and "off" states by guanine exchange factors. Although GDP release in monomeric GTPases is accelerated by guanine exchange factor-induced removal of magnesium from the catalytic site, we found that release of ADP from MutS is not influenced by the metal ion in this manner. Rather, ADP release is induced by the binding of mismatched DNA at the opposite end of the protein, a long-range allosteric response resembling the mechanism of activation of heterotrimeric GTPases. Magnesium influences switching in MutS by inducing faster and tighter ATP binding, allowing rapid downstream responses. MutS mutants with decreased affinity for the metal ion are impaired in fast switching and in vivo mismatch repair. Thus, the G-proteins and MutS conceptually employ the same efficient use of the high energy cofactor: slow hydrolysis in the absence of a signal and fast conversion to the active state when required.
- Division of Biochemistry and Center for Biomedical Genetics, Netherlands Cancer Institute, 1066 CX Amsterdam, The Netherlands. j.lebbink@erasmusmc.nl
Organizational Affiliation: