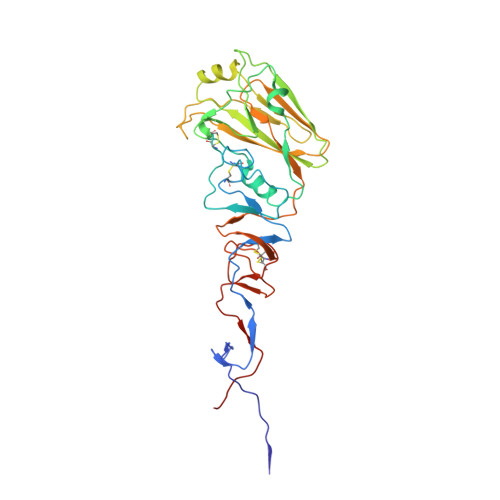
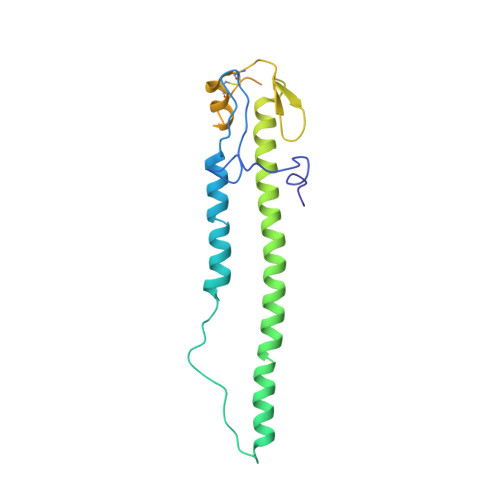
From the Cover: Structures of Receptor Complexes Formed by Hemagglutinins from the Asian Influenza Pandemic of 1957.
Liu, J., Stevens, D.J., Haire, L.F., Walker, P.A., Coombs, P.J., Russell, R.J., Gamblin, S.J., Skehel, J.J.(2009) Proc Natl Acad Sci U S A 106: 17175
- PubMed: 19805083
- DOI: https://doi.org/10.1073/pnas.0906849106
- Primary Citation of Related Structures:
2WR0, 2WR1, 2WR2, 2WR3, 2WR4, 2WR5, 2WR7, 2WRB, 2WRC, 2WRD, 2WRE, 2WRF, 2WRG, 2WRH - PubMed Abstract:
The viruses that caused the three influenza pandemics of the twentieth century in 1918, 1957, and 1968 had distinct hemagglutinin receptor binding glycoproteins that had evolved the capacity to recognize human cell receptors. We have determined the structure of the H2 hemagglutinin from the second pandemic, the "Asian Influenza" of 1957. We compare it with the 1918 "Spanish Influenza" hemagglutinin, H1, and the 1968 "Hong Kong Influenza" hemagglutinin, H3, and show that despite its close overall structural similarity to H1, and its more distant relationship to H3, the H2 receptor binding site is closely related to that of H3 hemagglutinin. By analyzing hemagglutinins of potential H2 avian precursors of the pandemic virus, we show that the human receptor can be bound by avian hemagglutinins that lack the human-specific mutations of H2 and H3 pandemic viruses, Gln-226Leu, and Gly-228Ser. We show how Gln-226 in the avian H2 receptor binding site, together with Asn-186, form hydrogen bond networks through bound water molecules to mediate binding to human receptor. We show that the human receptor adopts a very similar conformation in both human and avian hemagglutinin-receptor complexes. We also show that Leu-226 in the receptor binding site of human virus hemagglutinins creates a hydrophobic environment near the Sia-1-Gal-2 glycosidic linkage that favors binding of the human receptor and is unfavorable for avian receptor binding. We consider the significance for the development of pandemics, of the existence of avian viruses that can bind to both avian and human receptors.
- MRC National Institute for Medical Research, The Ridgeway, Mill Hill, London NW7 1AA, United Kingdom.
Organizational Affiliation: