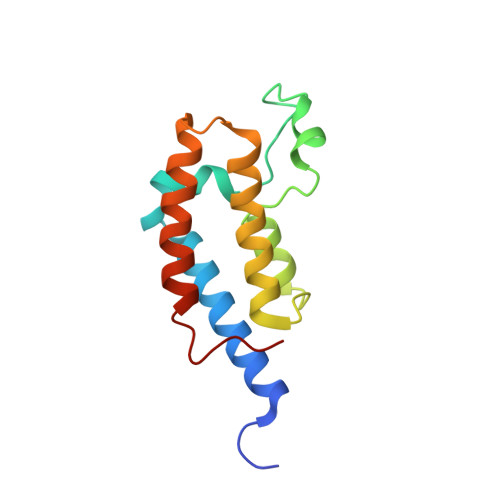
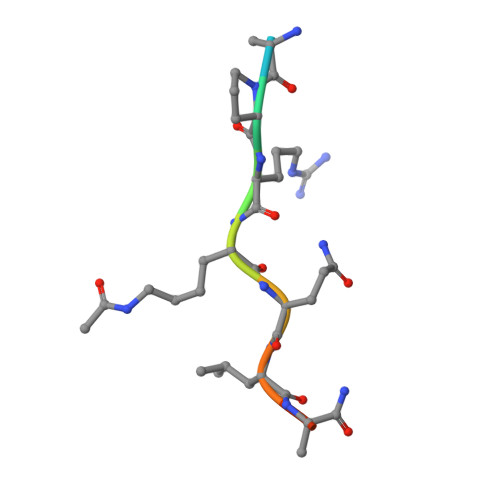
Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Moriniere, J., Rousseaux, S., Steuerwald, U., Soler-Lopez, M., Curtet, S., Vitte, A.-L., Govin, J., Gaucher, J., Sadoul, K., Hart, D.J., Krijgsveld, J., Khochbin, S., Mueller, C.W., Petosa, C.(2009) Nature 461: 664
- PubMed: 19794495
- DOI: https://doi.org/10.1038/nature08397
- Primary Citation of Related Structures:
2WP1, 2WP2 - PubMed Abstract:
A key step in many chromatin-related processes is the recognition of histone post-translational modifications by effector modules such as bromodomains and chromo-like domains of the Royal family. Whereas effector-mediated recognition of single post-translational modifications is well characterized, how the cell achieves combinatorial readout of histones bearing multiple modifications is poorly understood. One mechanism involves multivalent binding by linked effector modules. For example, the tandem bromodomains of human TATA-binding protein-associated factor-1 (TAF1) bind better to a diacetylated histone H4 tail than to monoacetylated tails, a cooperative effect attributed to each bromodomain engaging one acetyl-lysine mark. Here we report a distinct mechanism of combinatorial readout for the mouse TAF1 homologue Brdt, a testis-specific member of the BET protein family. Brdt associates with hyperacetylated histone H4 (ref. 7) and is implicated in the marked chromatin remodelling that follows histone hyperacetylation during spermiogenesis, the stage of spermatogenesis in which post-meiotic germ cells mature into fully differentiated sperm. Notably, we find that a single bromodomain (BD1) of Brdt is responsible for selectively recognizing histone H4 tails bearing two or more acetylation marks. The crystal structure of BD1 bound to a diacetylated H4 tail shows how two acetyl-lysine residues cooperate to interact with one binding pocket. Structure-based mutagenesis that reduces the selectivity of BD1 towards diacetylated tails destabilizes the association of Brdt with acetylated chromatin in vivo. Structural analysis suggests that other chromatin-associated proteins may be capable of a similar mode of ligand recognition, including yeast Bdf1, human TAF1 and human CBP/p300 (also known as CREBBP and EP300, respectively). Our findings describe a new mechanism for the combinatorial readout of histone modifications in which a single effector module engages two marks on a histone tail as a composite binding epitope.
- European Molecular Biology Laboratory, Grenoble Outstation, 6 rue Jules Horowitz, BP 181, 38042 Grenoble Cedex 9, France.
Organizational Affiliation: