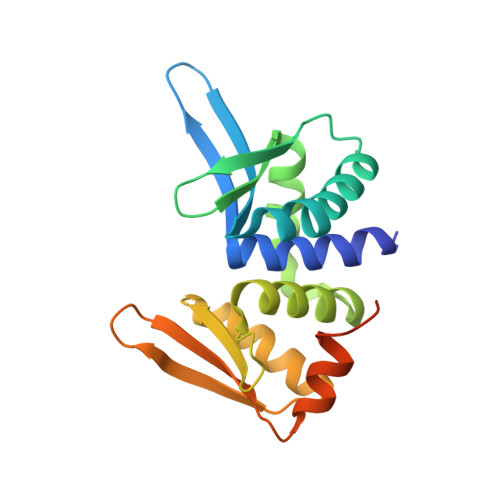
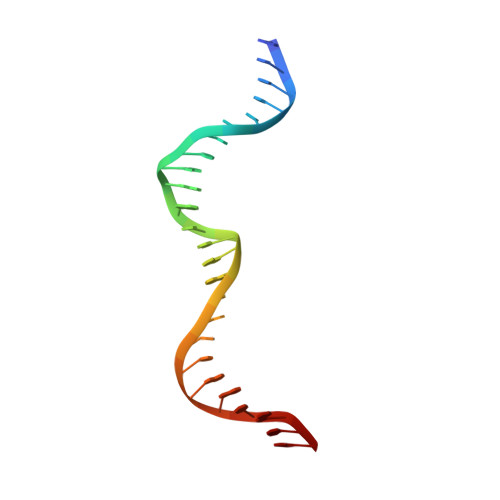
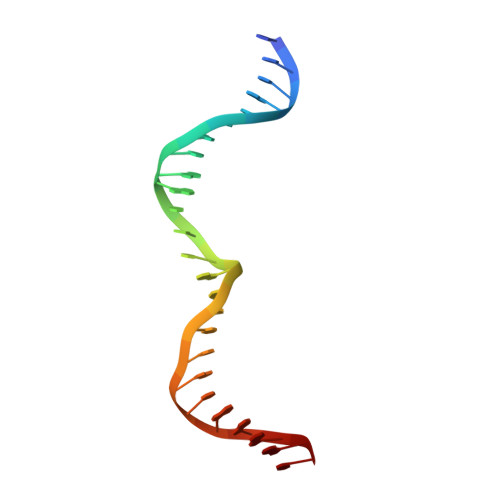
Crystal Structure of I-Dmoi in Complex with its Target DNA Provides New Insights Into Meganuclease Engineering.
Marcaida, M.J., Prieto, J., Redondo, P., Nadra, A.D., Alibes, A., Serrano, L., Grizot, S., Duchateau, P., Paques, F., Blanco, F.J., Montoya, G.(2008) Proc Natl Acad Sci U S A 105: 16888
- PubMed: 18974222
- DOI: https://doi.org/10.1073/pnas.0804795105
- Primary Citation of Related Structures:
2VS7, 2VS8 - PubMed Abstract:
Homing endonucleases, also known as meganucleases, are sequence-specific enzymes with large DNA recognition sites. These enzymes can be used to induce efficient homologous gene targeting in cells and plants, opening perspectives for genome engineering with applications in a wide series of fields, ranging from biotechnology to gene therapy. Here, we report the crystal structures at 2.0 and 2.1 A resolution of the I-DmoI meganuclease in complex with its substrate DNA before and after cleavage, providing snapshots of the catalytic process. Our study suggests that I-DmoI requires only 2 cations instead of 3 for DNA cleavage. The structure sheds light onto the basis of DNA binding, indicating key residues responsible for nonpalindromic target DNA recognition. In silico and in vivo analysis of the I-DmoI DNA cleavage specificity suggests that despite the relatively few protein-base contacts, I-DmoI is highly specific when compared with other meganucleases. Our data open the door toward the generation of custom endonucleases for targeted genome engineering using the monomeric I-DmoI scaffold.
- Macromolecular Crystallography and Nuclear Magnetic Resonance Groups, Structural Biology and Biocomputing Programme, Spanish National Cancer Research Centre, c/Melchor Fdez. Almagro 3, 28029 Madrid, Spain.
Organizational Affiliation: