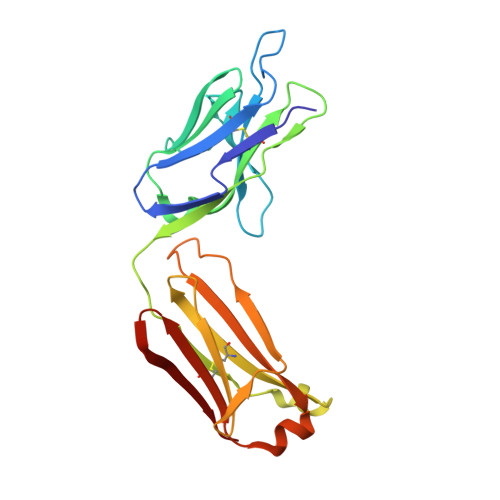
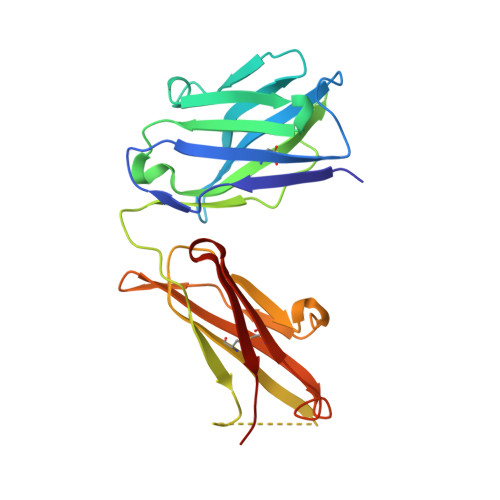
Characterization of a Diagnostic Fab Fragment Binding Trimeric Lewis X.
De Geus, D.C., Van Roon, A.M.M., Thomassen, E.A.J., Hokke, C.H., Deelder, A.M., Abrahams, J.P.(2009) Proteins 76: 439
- PubMed: 19173313
- DOI: https://doi.org/10.1002/prot.22356
- Primary Citation of Related Structures:
2VQ1 - PubMed Abstract:
Lewis X trisaccharides normally function as essential cell-cell interaction mediators. However, oligomers of Lewis X trisaccharides expressed by the parasite Schistosoma mansoni seem to be related to its evasion of the immune response of its human host. Here we show that monoclonal antibody 54-5C10-A, which is used to diagnose schistosomiasis in humans, interacts with oligomers of at least three Lewis X trisaccharides, but not with monomeric Lewis X. We describe the sequence and the 2.5 A crystal structure of its Fab fragment and infer a possible mode of binding of the polymeric Lewis X from docking studies. Our studies indicate a radically different mode of binding compared to Fab 291-2G3-A, which is specific for monomeric Lewis X, thus providing a structural explanation of the diagnostic success of 54-5C10-A.
- Leiden University, The Netherlands. d.de.geus@chem.leidenuniv.nl
Organizational Affiliation: