X-Ray Structure of the Phf Core C-Terminus: Insight Into the Folding of the Intrinsically Disordered Protein Tau in Alzheimer'S Disease.
Sevcik, J., Skrabana, R., Dvorsky, R., Csokova, N., Iqbal, K., Novak, M.(2007) FEBS Lett 581: 5872
- PubMed: 18061582
- DOI: https://doi.org/10.1016/j.febslet.2007.11.067
- Primary Citation of Related Structures:
2V17 - PubMed Abstract:
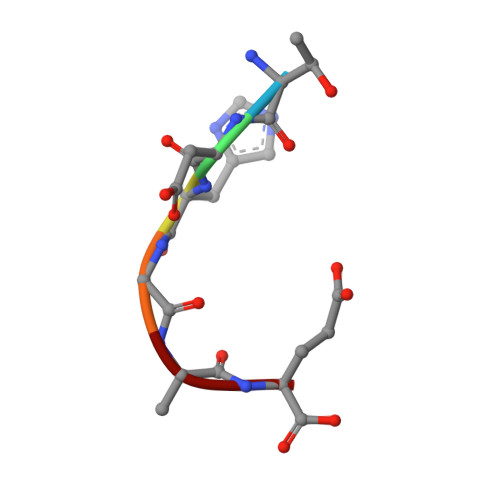
The major constituent of Alzheimer's disease paired helical filaments (PHF) core is intrinsically disordered protein (IDP) tau. In spite of a considerable effort, insoluble character of PHF together with inherent physical properties of IDP tau have precluded so far reconstruction of PHF 3D structure by X-ray crystallography or NMR spectroscopy. Here we present first crystallographic study of PHF core C-terminus. Using monoclonal antibody MN423 specific to the tertiary structure of the PHF core, the in vivo PHF structure was imprinted into recombinant core PHF tau. Crystallization of the complex led to determination of the structure of the core PHF tau protein fragment 386TDHGAE391 at 1.65A resolution. Structural analysis suggests important role of the core PHF C-terminus for PHF assembly. It is reasonable to expect that this approach will help to reveal the structural principles underlying the tau protein assembly into PHF and possibly will facilitate rationale drug design for inhibition of Alzheimer neurofibrillary changes.
- Institute of Neuroimmunology, Slovak Academy of Sciences, Dubravska cesta 9, 845 10 Bratislava, Slovakia.
Organizational Affiliation: