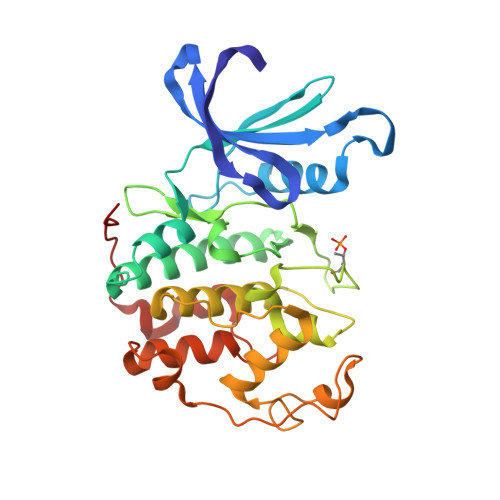
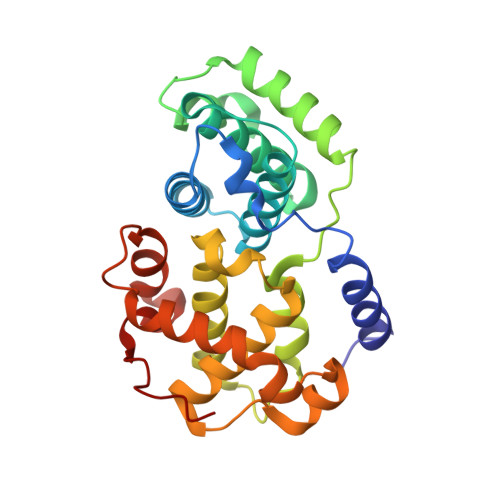
Discovery of a Potent Cdk2 Inhibitor with a Novel Binding Mode, Using Virtual Screening and Initial, Structure-Guided Lead Scoping.
Richardson, C.M., Nunns, C.L., Williamson, D.S., Parratt, M.J., Dokurno, P., Howes, R., Borgognoni, J., Drysdale, M.J., Finch, H., Hubbard, R.E., Jackson, P.S., Kierstan, P., Lentzen, G., Moore, J.D., Murray, J.B., Simmonite, H., Surgenor, A.E., Torrance, C.J.(2007) Bioorg Med Chem Lett 17: 3880
- PubMed: 17570665
- DOI: https://doi.org/10.1016/j.bmcl.2007.04.110
- Primary Citation of Related Structures:
2UZB, 2UZD, 2UZE, 2UZL, 2UZN, 2UZO, 2V0D - PubMed Abstract:
Virtual screening against a pCDK2/cyclin A crystal structure led to the identification of a potent and novel CDK2 inhibitor, which exhibited an unusual mode of interaction with the kinase binding motif. With the aid of X-ray crystallography and modelling, a medicinal chemistry strategy was implemented to probe the interactions seen in the crystal structure and to establish SAR. A fragment-based approach was also considered but a different, more conventional, binding mode was observed. Compound selectivity against GSK-3beta was improved using a rational design strategy, with crystallographic verification of the CDK2 binding mode.
- Vernalis (R&D) Ltd, Granta Park, Cambridge CB21 6GB, UK. c.richardson@vernalis.com
Organizational Affiliation: