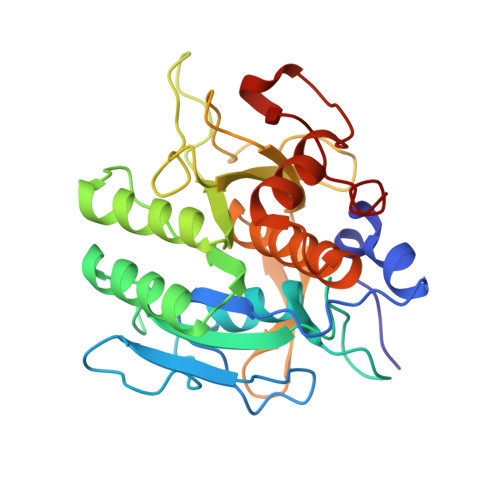
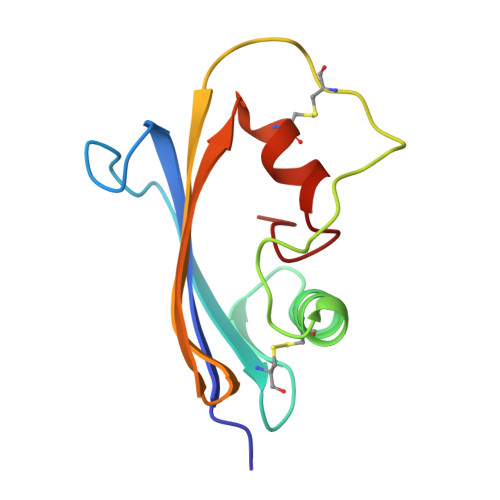
Refined crystal structure of the complex of subtilisin BPN' and Streptomyces subtilisin inhibitor at 1.8 A resolution.
Takeuchi, Y., Satow, Y., Nakamura, K.T., Mitsui, Y.(1991) J Mol Biology 221: 309-325
- PubMed: 1920411
- Primary Citation of Related Structures:
2SIC - PubMed Abstract:
The crystal structure of subtilisin BPN' complexed with a proteinaceous inhibitor SSI (Streptomyces subtilisin inhibitor) was refined at 1.8 A resolution to an R-factor of 0.177 with a root-mean-square deviation from ideal bond lengths of 0.014 A. The work finally established that the SSI-subtilisin complex is a Michaelis complex with a distance between the O gamma of active Ser221 and the carbonyl carbon of the scissile peptide bond being an intermediate value between a covalent bond and a van der Waals' contact, 2.7 A. This feature, as well as the geometry of the catalytic triad and the oxyanion hole, is coincident with that found in other highly refined crystal structures of the complex of subtilisin Novo, subtilisin Carlsberg, bovine trypsin or Streptomyces griseus protease B with their proteinaceous inhibitors. The enzyme-inhibitor beta-sheet interaction is composed of two separate parts: that between the P1-P3 residues of SSI and the 125-127 chain segment (the "S1-3 site") of subtilisin and that between the P4-P6 residues of SSI and th 102-104 chain segment (the "S4-6 site") of subtilisin. The latter beta-interaction is unique to subtilisin. In contrast, the beta-sheet interaction previously found in the complex of subtilisin Novo and chymotrypsin inhibitor 2 or in the complex of subtilisin Carlsberg and Eglin C is distinct from the present complex in that the two types of beta-interactions are not separate. As for the flexibility of the molecules comprising the present complex, the following observations were made by comparing the B-factors for free and complexed SSI and comparing those for free and complexed subtilisin BPN'. The rigidification of the component molecules upon complex formation occurs in a very localized region: in SSI, the "primary" and "secondary" contact regions and the flanking region; in subtilisin BPN', the S1-3 and S4-6 sites and the flanking region.
- Pharmaceutical Research Center, Meiji Seika Kaisha, Ltd., Yokohama, Japan.
Organizational Affiliation: