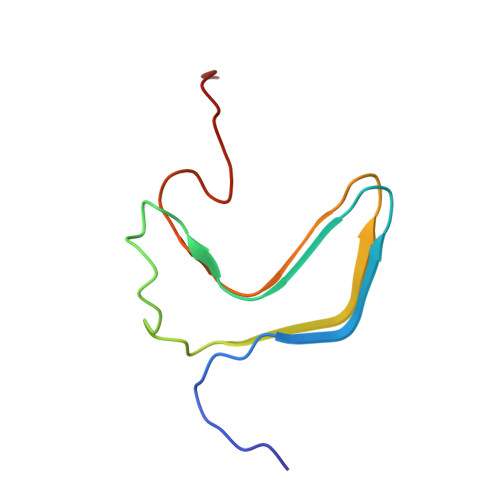
Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core
Wasmer, C., Lange, A., Van Melckebeke, H., Siemer, A.B., Riek, R., Meier, B.H.(2008) Science 319: 1523-1526
- PubMed: 18339938
- DOI: https://doi.org/10.1126/science.1151839
- Primary Citation of Related Structures:
2RNM - PubMed Abstract:
Prion and nonprion forms of proteins are believed to differ solely in their three-dimensional structure, which is therefore of paramount importance for the prion function. However, no atomic-resolution structure of the fibrillar state that is likely infectious has been reported to date. We present a structural model based on solid-state nuclear magnetic resonance restraints for amyloid fibrils from the prion-forming domain (residues 218 to 289) of the HET-s protein from the filamentous fungus Podospora anserina. On the basis of 134 intra- and intermolecular experimental distance restraints, we find that HET-s(218-289) forms a left-handed beta solenoid, with each molecule forming two helical windings, a compact hydrophobic core, at least 23 hydrogen bonds, three salt bridges, and two asparagine ladders. The structure is likely to have broad implications for understanding the infectious amyloid state.
- Physical Chemistry, ETH Zurich, 8093 Zurich, Switzerland.
Organizational Affiliation: