Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4.
Wong, J.H., Fiala, K.A., Suo, Z., Ling, H.(2008) J Mol Biology 379: 317-330
- PubMed: 18448122
- DOI: https://doi.org/10.1016/j.jmb.2008.03.038
- Primary Citation of Related Structures:
2RDI, 2RDJ - PubMed Abstract:
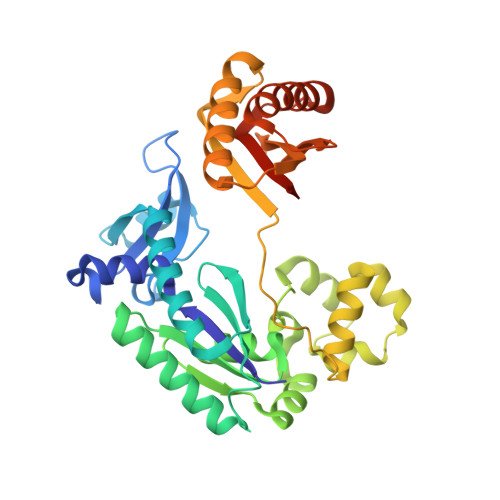
Y-family DNA polymerases catalyze translesion DNA synthesis over damaged DNA. Each Y-family polymerase has a polymerase core consisting of a palm, finger and thumb domain in addition to a fourth domain known as a little finger domain. It is unclear how each domain moves during nucleotide incorporation and what type of conformational changes corresponds to the rate-limiting step previously reported in kinetic studies. Here, we present three crystal structures of the prototype Y-family polymerase: apo-Dpo4 at 1.9 A resolution, Dpo4-DNA binary complex and Dpo4-DNA-dTMP ternary complex at 2.2 A resolution. Dpo4 undergoes dramatic conformational changes from the apo to the binary structures with a 131 degrees rotation of the little finger domain relative to the polymerase core upon DNA binding. This DNA-induced conformational change is verified in solution by our tryptophan fluorescence studies. In contrast, the polymerase core retains the same conformation in all three conformationally distinct states. Particularly, the finger domain which is responsible for checking base pairing between the template base and an incoming nucleotide retains a rigid conformation. The inflexibility of the polymerase core likely contributes to the low fidelity of Dpo4, in addition to its loose and solvent-accessible active site. Interestingly, while the binary and ternary complexes of Dpo4 retain an identical global conformation, the aromatic side chains of two conserved tyrosines at the nucleotide-binding site change orientations between the binary and ternary structures. Such local conformational changes may correspond to the rate-limiting step in the mechanism of nucleotide incorporation. Together, the global and local conformational transitions observed in our study provide a structural basis for the distinct kinetic steps of a catalytic cycle of DNA polymerization performed by a Y-family polymerase.
- Department of Biochemistry, University of Western Ontario, London, Ontario, Canada.
Organizational Affiliation: