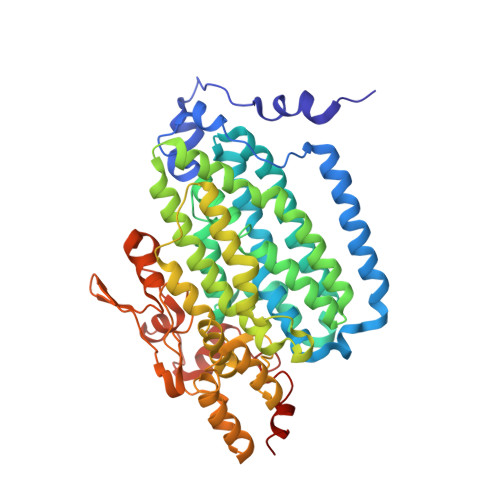
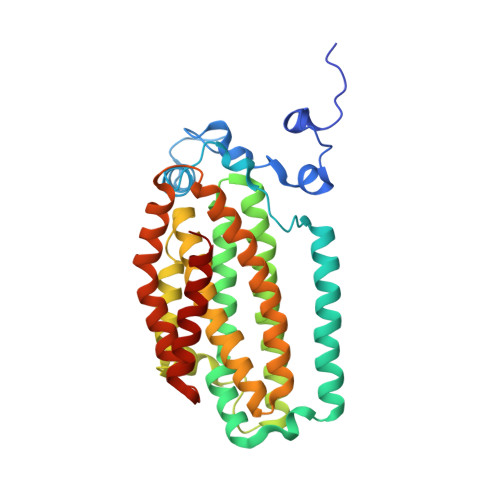
Dioxygen activation at non-heme diiron centers: oxidation of a proximal residue in the I100W variant of toluene/o-xylene monooxygenase hydroxylase.
Murray, L.J., Garcia-Serres, R., McCormick, M.S., Davydov, R., Naik, S.G., Kim, S.H., Hoffman, B.M., Huynh, B.H., Lippard, S.J.(2007) Biochemistry 46: 14795-14809
- PubMed: 18044971
- DOI: https://doi.org/10.1021/bi7017128
- Primary Citation of Related Structures:
2RDB - PubMed Abstract:
At its carboxylate-bridged diiron active site, the hydroxylase component of toluene/o-xylene monooxygenase activates dioxygen for subsequent arene hydroxylation. In an I100W variant of this enzyme, we characterized the formation and decay of two species formed by addition of dioxygen to the reduced, diiron(II) state by rapid-freeze quench (RFQ) EPR, Mössbauer, and ENDOR spectroscopy. The dependence of the formation and decay rates of this mixed-valent transient on pH and the presence of phenol, propylene, or acetylene was investigated by double-mixing stopped-flow optical spectroscopy. Modification of the alpha-subunit of the hydroxylase after reaction of the reduced protein with dioxygen-saturated buffer was investigated by tryptic digestion coupled mass spectrometry. From these investigations, we conclude that (i) a diiron(III,IV)-W* transient, kinetically linked to a preceding diiron(III) intermediate, arises from the one-electron oxidation of W100, (ii) the tryptophan radical is deprotonated, (iii) rapid exchange of either a terminal water or hydroxide ion with water occurs at the ferric ion in the diiron(III,IV) cluster, and (iv) the diiron(III,IV) core and W* decay to the diiron(III) product by a common mechanism. No transient radical was observed by stopped-flow optical spectroscopy for reactions of the reduced hydroxylase variants I100Y, L208F, and F205W with dioxygen. The absence of such species, and the deprotonated state of the tryptophanyl radical in the diiron(III,IV)-W* transient, allow for a conservative estimate of the reduction potential of the diiron(III) intermediate as lying between 1.1 and 1.3 V. We also describe the X-ray crystal structure of the I100W variant of ToMOH.
- Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA.
Organizational Affiliation: