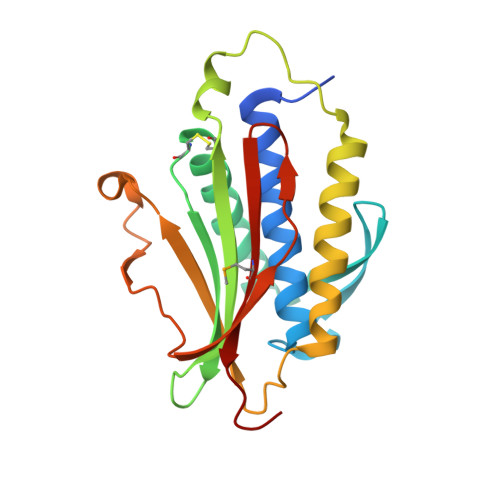
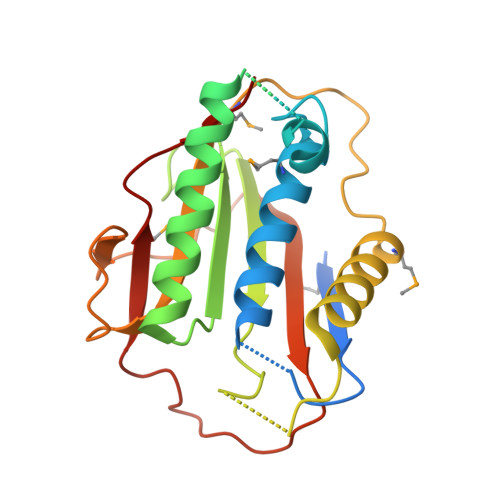
p31comet blocks Mad2 activation through structural mimicry.
Yang, M., Li, B., Tomchick, D.R., Machius, M., Rizo, J., Yu, H., Luo, X.(2007) Cell 131: 744-755
- PubMed: 18022368
- DOI: https://doi.org/10.1016/j.cell.2007.08.048
- Primary Citation of Related Structures:
2QYF - PubMed Abstract:
The status of spindle checkpoint signaling depends on the balance of two opposing dynamic processes that regulate the highly unusual two-state behavior of Mad2. In mitosis, a Mad1-Mad2 core complex recruits cytosolic Mad2 to kinetochores through Mad2 dimerization and converts Mad2 to a conformer amenable to Cdc20 binding, thereby facilitating checkpoint activation. p31(comet) inactivates the checkpoint through binding to Mad1- or Cdc20-bound Mad2, thereby preventing Mad2 activation and promoting the dissociation of the Mad2-Cdc20 complex. Here, we report the crystal structure of the Mad2-p31(comet) complex. The C-terminal region of Mad2 that undergoes rearrangement in different Mad2 conformers is a major structural determinant for p31(comet) binding, explaining the specificity of p31(comet) toward Mad1- or Cdc20-bound Mad2. p31(comet) adopts a fold strikingly similar to that of Mad2 and binds at the dimerization interface of Mad2. Thus, p31(comet) exploits the two-state behavior of Mad2 to block its activation by acting as an "anti-Mad2."
- Department of Pharmacology, The University of Texas Southwestern Medical Center, 6001 Forest Park Road, Dallas, TX 75390, USA.
Organizational Affiliation: