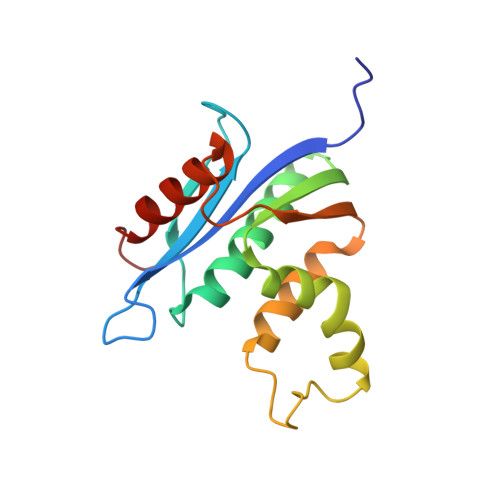
Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Nowotny, M., Gaidamakov, S.A., Ghirlando, R., Cerritelli, S.M., Crouch, R.J., Yang, W.(2007) Mol Cell 28: 264-276
- PubMed: 17964265
- DOI: https://doi.org/10.1016/j.molcel.2007.08.015
- Primary Citation of Related Structures:
2QK9, 2QKB, 2QKK - PubMed Abstract:
We report here crystal structures of human RNase H1 complexed with an RNA/DNA substrate. Unlike B. halodurans RNase H1, human RNase H1 has a basic protrusion, which forms a DNA-binding channel and together with the conserved phosphate-binding pocket confers specificity for the B form and 2'-deoxy DNA. The RNA strand is recognized by four consecutive 2'-OH groups and cleaved by a two-metal ion mechanism. Although RNase H1 is overall positively charged, the substrate interface is neutral to acidic in character, which likely contributes to the catalytic specificity. Positions of the scissile phosphate and two catalytic metal ions are interdependent and highly coupled. Modeling of HIV reverse transcriptase (RT) with RNA/DNA in its RNase H active site suggests that the substrate cannot simultaneously occupy the polymerase active site and must undergo a conformational change to toggle between the two catalytic centers. The region that accommodates this conformational change offers a target to develop HIV-specific inhibitors.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: