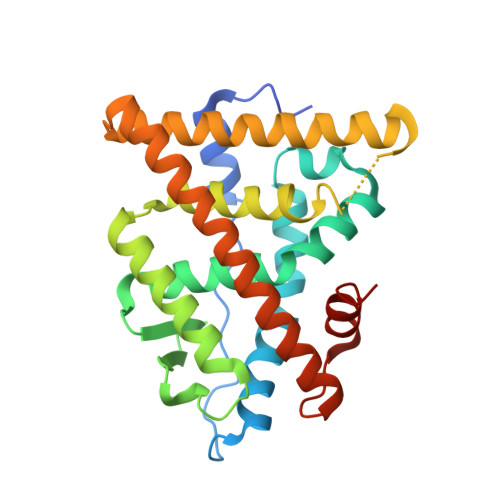
Elemental isomerism: a boron-nitrogen surrogate for a carbon-carbon double bond increases the chemical diversity of estrogen receptor ligands
Zhou, H.-B., Nettles, K.W., Bruning, J.B., Kim, Y., Joachimiak, A., Sharma, S., Carlson, K.E., Stossi, F., Katzenellenbogen, B.S., Greene, G.L., Katzenellenbogen, J.A.(2007) Chem Biol 14: 659-669
- PubMed: 17584613
- DOI: https://doi.org/10.1016/j.chembiol.2007.04.009
- Primary Citation of Related Structures:
2Q6J - PubMed Abstract:
To increase the chemical diversity of bioactive molecules by incorporating unusual elements, we have examined the replacement of a C=C double bond with the isoelectronic, isostructural B-N bond in the context of nonsteroidal estrogen receptor (ER) ligands. While the B-N bond was hydrolytically labile in the unhindered cyclofenil system, the more hindered anilino dimesitylboranes, analogs of triarylethylene estrogens, were easily prepared, hydrolytically stable, and demonstrated substantial affinity for ERs. X-ray analysis of one ERalpha-ligand complex revealed steric clashes with the para methyl groups distorting the receptor; removal of these groups resulted in an increase in affinity, potency, and transcriptional efficacy. These studies define the structural determinants of stability and cellular bioactivity of a B-N for C=C substitution in nonsteroidal estrogens and provide a framework for further exploration of "elemental isomerism" for diversification of drug-like molecules.
- Department of Chemistry, University of Illinois, Urbana, IL 61801, USA.
Organizational Affiliation: