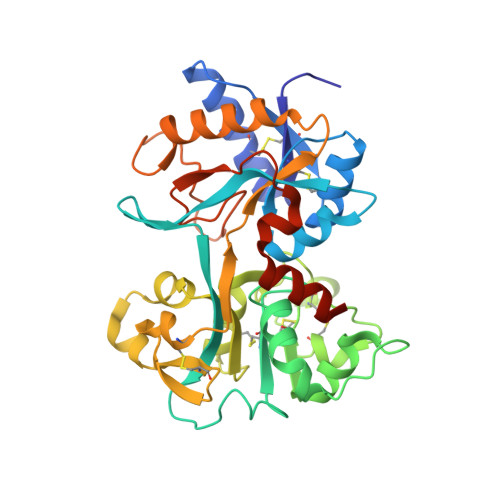
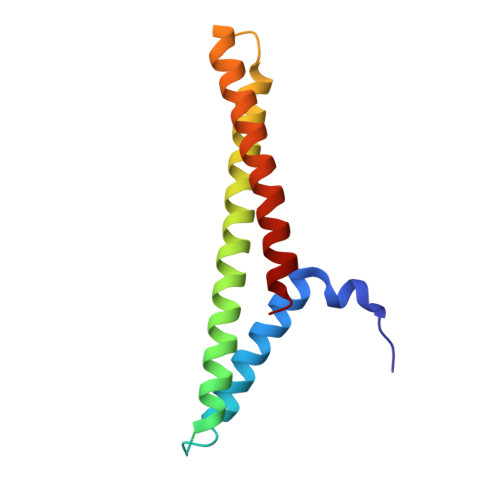
Structure of a Complex of Human Lactoferrin N-lobe with Pneumococcal Surface Protein A Provides Insight into Microbial Defense Mechanism.
Senkovich, O., Cook, W.J., Mirza, S., Hollingshead, S.K., Protasevich, I.I., Briles, D.E., Chattopadhyay, D.(2007) J Mol Biology 370: 701-713
- PubMed: 17543335
- DOI: https://doi.org/10.1016/j.jmb.2007.04.075
- Primary Citation of Related Structures:
2PMS - PubMed Abstract:
Human lactoferrin, a component of the innate immune system, kills a wide variety of microorganisms including the Gram positive bacteria Streptococcus pneumoniae. Pneumococcal surface protein A (PspA) efficiently inhibits this bactericidal action. The crystal structure of a complex of the lactoferrin-binding domain of PspA with the N-lobe of human lactoferrin reveals direct and specific interactions between the negatively charged surface of PspA helices and the highly cationic lactoferricin moiety of lactoferrin. Binding of PspA blocks surface accessibility of this bactericidal peptide preventing it from penetrating the bacterial membrane. Results of site-directed mutagenesis, in vitro protein binding assays and isothermal titration calorimetry measurements corroborate that the specific electrostatic interactions observed in the crystal structure represent major associations between PspA and lactoferrin. The structure provides a snapshot of the protective mechanism utilized by pathogens against the host's first line of defense. PspA represents a major virulence factor and a promising vaccine candidate. Insights from the structure of the complex have implications for designing therapeutic strategies for treatment and prevention of pneumococcal diseases that remain a major public health problem worldwide.
- Center for Biophysical Sciences and Engineering, University of Alabama at Birmingham, AL 35294, USA.
Organizational Affiliation: