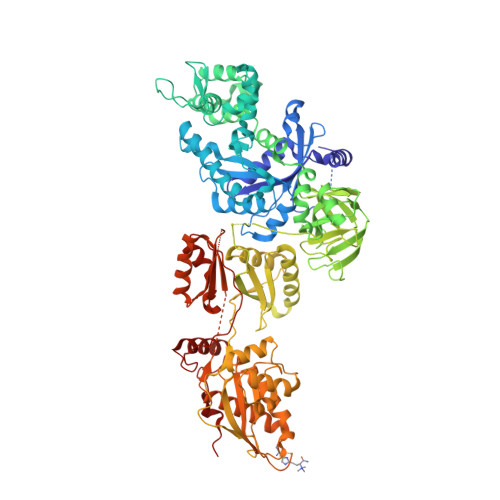
Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Taylor, D.J., Nilsson, J., Merrill, A.R., Andersen, G.R., Nissen, P., Frank, J.(2007) EMBO J 26: 2421-2431
- PubMed: 17446867
- DOI: https://doi.org/10.1038/sj.emboj.7601677
- Primary Citation of Related Structures:
2P8W, 2P8X, 2P8Y, 2P8Z - PubMed Abstract:
On the basis of kinetic data on ribosome protein synthesis, the mechanical energy for translocation of the mRNA-tRNA complex is thought to be provided by GTP hydrolysis of an elongation factor (eEF2 in eukaryotes, EF-G in bacteria). We have obtained cryo-EM reconstructions of eukaryotic ribosomes complexed with ADP-ribosylated eEF2 (ADPR-eEF2), before and after GTP hydrolysis, providing a structural basis for analyzing the GTPase-coupled mechanism of translocation. Using the ADP-ribosyl group as a distinct marker, we observe conformational changes of ADPR-eEF2 that are due strictly to GTP hydrolysis. These movements are likely representative of native eEF2 motions in a physiological context and are sufficient to uncouple the mRNA-tRNA complex from two universally conserved bases in the ribosomal decoding center (A1492 and A1493 in Escherichia coli) during translocation. Interpretation of these data provides a detailed two-step model of translocation that begins with the eEF2/EF-G binding-induced ratcheting motion of the small ribosomal subunit. GTP hydrolysis then uncouples the mRNA-tRNA complex from the decoding center so translocation of the mRNA-tRNA moiety may be completed by a head rotation of the small subunit.
- Howard Hughes Medical Institute, Health Research Inc.,Wadsworth Center, Albany, NY 12201-0509, USA.
Organizational Affiliation: