Low resolution solution structure of the 152 kDa complex between nitrite reductase and pseudoazurin from A. faecalis by paramagnetic NMR.
Vlasie, M.D., Fernandez-Busnadiego, R., Prudencio, M., Ubbink, M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Copper-containing nitrite reductase | 341 | Alcaligenes faecalis | Mutation(s): 0 Gene Names: nirK, nir EC: 1.7.2.1 | 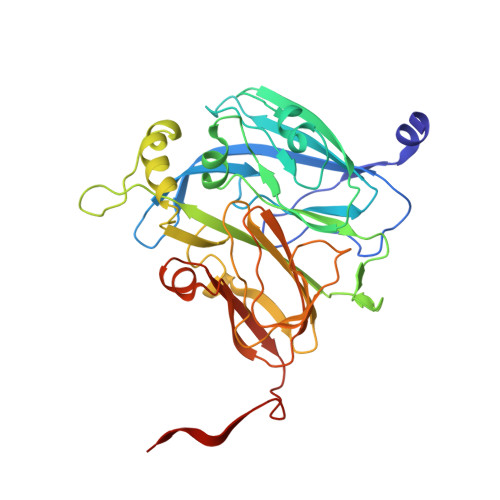 | |
UniProt | |||||
Find proteins for P38501 (Alcaligenes faecalis) Explore P38501 Go to UniProtKB: P38501 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P38501 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Pseudoazurin | 123 | Alcaligenes faecalis | Mutation(s): 0 | 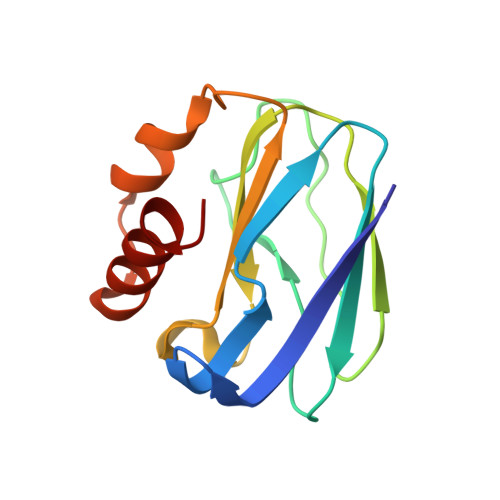 | |
UniProt | |||||
Find proteins for P04377 (Alcaligenes faecalis) Explore P04377 Go to UniProtKB: P04377 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04377 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GD Query on GD | G [auth A] H [auth A] I [auth A] L [auth B] M [auth B] | GADOLINIUM ATOM Gd UIWYJDYFSGRHKR-UHFFFAOYSA-N |  | ||
| CU Query on CU | E [auth A] F [auth A] J [auth B] K [auth B] O [auth C] | COPPER (II) ION Cu JPVYNHNXODAKFH-UHFFFAOYSA-N |  | ||