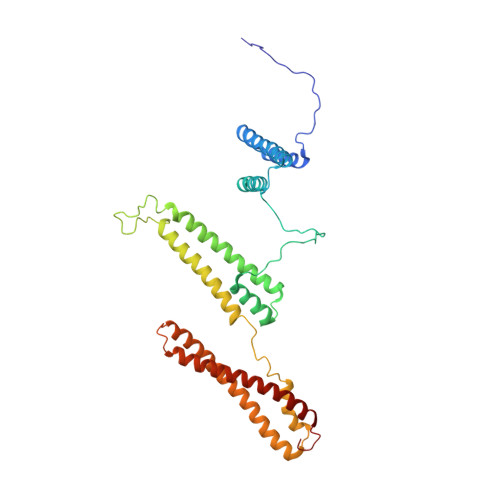
The structure of receptor-associated protein (RAP).
Lee, D., Walsh, J.D., Migliorini, M., Yu, P., Cai, T., Schwieters, C.D., Krueger, S., Strickland, D.K., Wang, Y.X.(2007) Protein Sci 16: 1628-1640
- PubMed: 17656581
- DOI: https://doi.org/10.1110/ps.072865407
- Primary Citation of Related Structures:
2P01, 2P03 - PubMed Abstract:
The receptor-associated protein (RAP) is a molecular chaperone that binds tightly to certain newly synthesized LDL receptor family members in the endoplasmic reticulum (ER) and facilitates their delivery to the Golgi. We have adopted a divide-and-conquer strategy to solve the structures of the individual domains of RAP using NMR spectroscopy. We present here the newly determined structure of domain 2. Based on this structure and the structures of domains 1 and 3, which were solved previously, we utilized experimental small-angle neutron scattering (SANS) data and a novel simulated annealing protocol to characterize the overall structure of RAP. The results reveal that RAP adopts a unique structural architecture consisting of three independent three-helix bundles that are connected by long and flexible linkers. The flexible linkers and the quasi-repetitive structural architecture may allow RAP to adopt various possible conformations when interacting with the LDL receptors, which are also made of repetitive substructure units.
- Protein-Nucleic Acid Interaction Section, Structural Biophysics Laboratory, National Cancer Institute at Frederick, National Institutes of Health, Frederick, Maryland 21702, USA.
Organizational Affiliation: