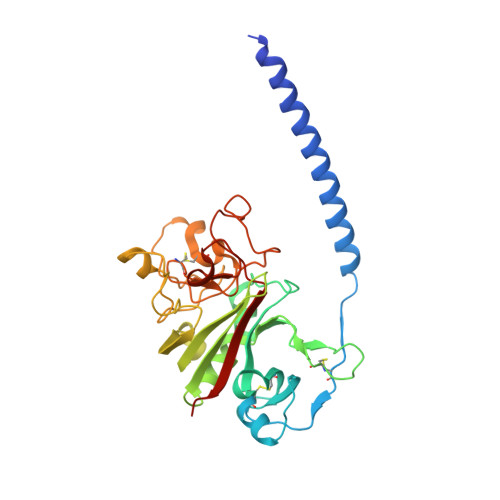
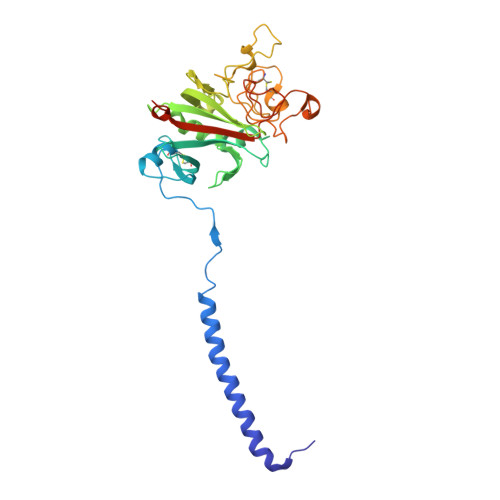
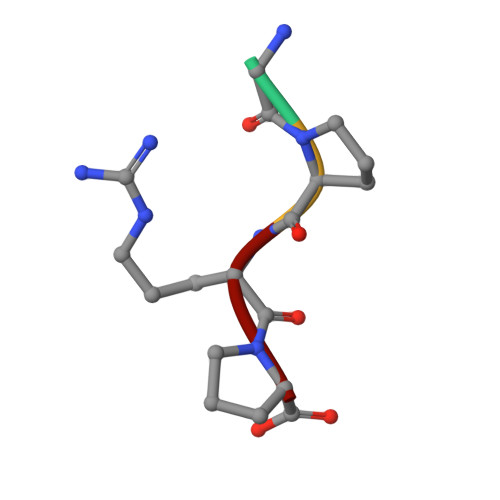
Probing the gamma2 Calcium-Binding Site: Studies with gammaD298,301A Fibrinogen Reveal Changes in the gamma294-301 Loop that Alter the Integrity of the "a" Polymerization Site.
Kostelansky, M.S., Lounes, K.C., Ping, L.F., Dickerson, S.K., Gorkun, O.V., Lord, S.T.(2007) Biochemistry 46: 5114-5123
- PubMed: 17411074
- DOI: https://doi.org/10.1021/bi602607a
- Primary Citation of Related Structures:
2OYH, 2OYI - PubMed Abstract:
To determine the significance of the gamma2 calcium-binding site in fibrin polymerization, we synthesized the fibrinogen variant, gammaD298,301A. We expected these two alanine substitutions to prevent calcium binding in the gamma2 site. We examined the influence of calcium on the polymerization of gammaD298,301A fibrinogen, evaluated its plasmin susceptibility, and solved 2.7 and 2.4 A crystal structures of the variant with the peptide ligands Gly-Pro-Arg-Pro-amide (GPRP) and Gly-His-Arg-Pro-amide (GHRP), respectively. We found that thrombin-catalyzed polymerization of gammaD298,301A fibrinogen was modestly impaired, whereas batroxobin-catalyzed polymerization was significantly impaired relative to normal fibrinogen. Notably, the influence of calcium on polymerization was the same for the variant and for normal fibrinogen. Fibrinogen gammaD298,301A was more susceptible to plasmin proteolysis in the presence of GPRP. This finding suggests structural changes in the near-by "a" polymerization site. Comparisons of the structures revealed minor conformational changes in the gamma294-301 loop that are likely responsible for the weakened "a" site. When considered altogether, the data suggest that the gamma2 calcium-binding site does not significantly modulate polymerization. We cannot, however, rule out the possibility that the weakened "a" polymerization site masks an important role for the gamma2 calcium-binding site in normal polymerization. Somewhat unexpectedly, the structure data showed that GPRP bound to the "b" site and induced the same local conformational changes as GHRP to this site. This structure shows that "A:b" interactions can occur and suggests that these may participate in normal polymerization.
- Department of Chemistry, University of North Carolina, Chapel Hill, North Carolina 27599, USA.
Organizational Affiliation: