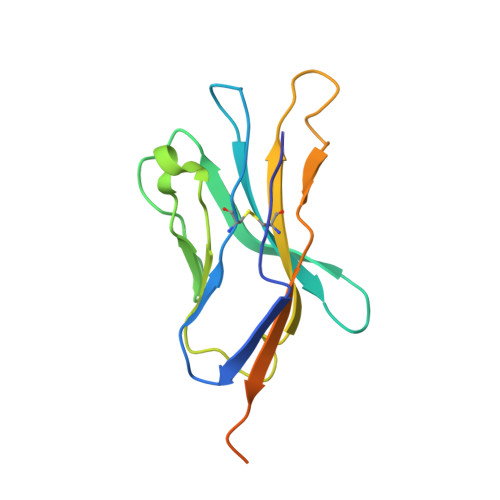
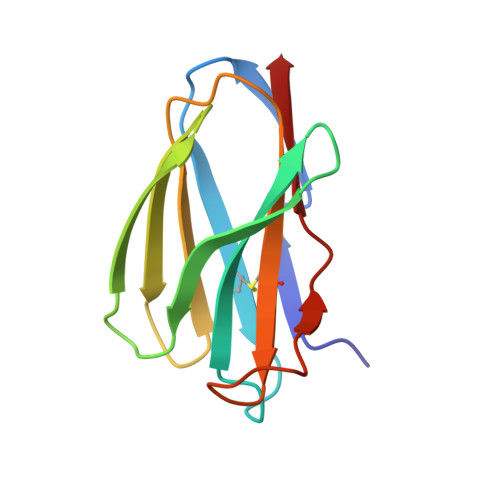
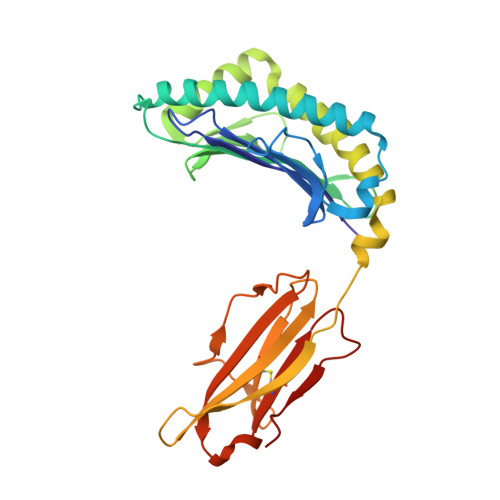
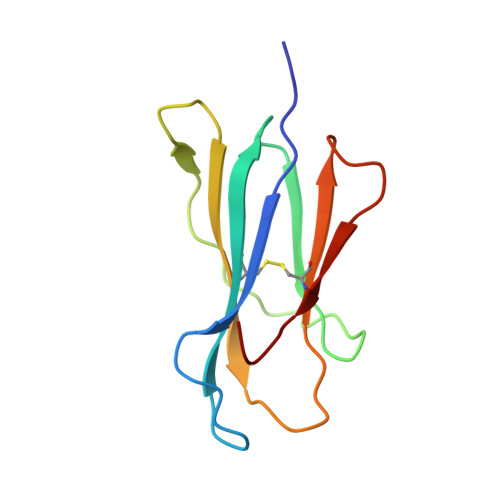
How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides?
Mazza, C., Auphan-Anezin, N., Gregoire, C., Guimezanes, A., Kellenberger, C., Roussel, A., Kearney, A., van der Merwe, P.A., Schmitt-Verhulst, A.M., Malissen, B.(2007) EMBO J 26: 1972-1983
- PubMed: 17363906
- DOI: https://doi.org/10.1038/sj.emboj.7601605
- Primary Citation of Related Structures:
2OL3 - PubMed Abstract:
Binding degeneracy is thought to constitute a fundamental property of the T-cell antigen receptor (TCR), yet its structural basis is poorly understood. We determined the crystal structure of a complex involving the BM3.3 TCR and a peptide (pBM8) bound to the H-2K(bm8) major histocompatibility complex (MHC) molecule, and compared it with the structures of the BM3.3 TCR bound to H-2K(b) molecules loaded with two peptides that had a minimal level of primary sequence identity with pBM8. Our findings provide a refined structural view of the basis of BM3.3 TCR cross-reactivity and a structural explanation for the long-standing paradox that a TCR antigen-binding site can be both specific and degenerate. We also measured the thermodynamic features and biological penalties that incurred during cross-recognition. Our data illustrate the difficulty for a given TCR in adapting to distinct peptide-MHC surfaces while still maintaining affinities that result in functional in vivo responses. Therefore, when induction of protective effector T cells is used as the ultimate criteria for adaptive immunity, TCRs are probably much less degenerate than initially assumed.
- Centre d'Immunologie de Marseille-Luminy, Université de la Méditerrannée, 13288 Marseille Cedex 09, France.
Organizational Affiliation: