Structural basis for viral late-domain binding to Alix
Lee, S., Joshi, A., Nagashima, K., Freed, E.O., Hurley, J.H.(2007) Nat Struct Mol Biol 14: 194-199
- PubMed: 17277784
- DOI: https://doi.org/10.1038/nsmb1203
- Primary Citation of Related Structures:
2OJQ - PubMed Abstract:
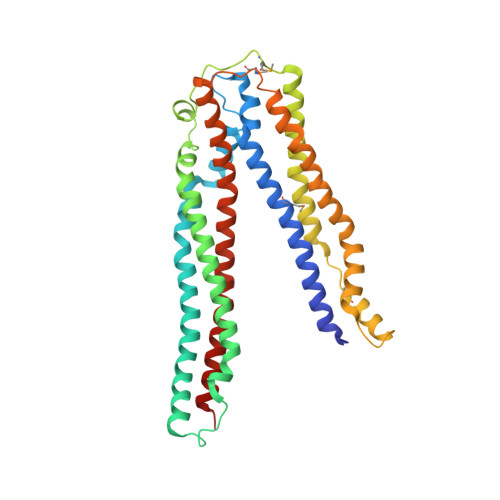
The modular protein Alix is a central node in endosomal-lysosomal trafficking and the budding of human immunodeficiency virus (HIV)-1. The Gag p6 protein of HIV-1 contains a LYPx(n)LxxL motif that is required for Alix-mediated budding and binds a region of Alix spanning residues 360-702. The structure of this fragment of Alix has the shape of the letter 'V' and is termed the V domain. The V domain has a topologically complex arrangement of 11 alpha-helices, with connecting loops that cross three times between the two arms of the V. The conserved residue Phe676 is at the center of a large hydrophobic pocket and is crucial for binding to a peptide model of HIV-1 p6. Overexpression of the V domain inhibits HIV-1 release from cells. This inhibition of release is reversed by mutations that block binding of the Alix V domain to p6.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH), US Department of Health and Human Services, Bethesda, Maryland 20892, USA.
Organizational Affiliation: