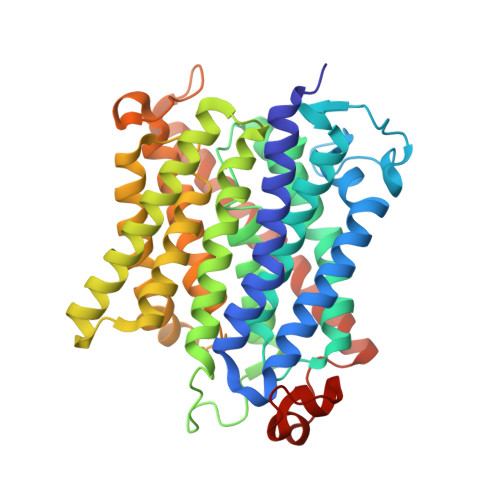
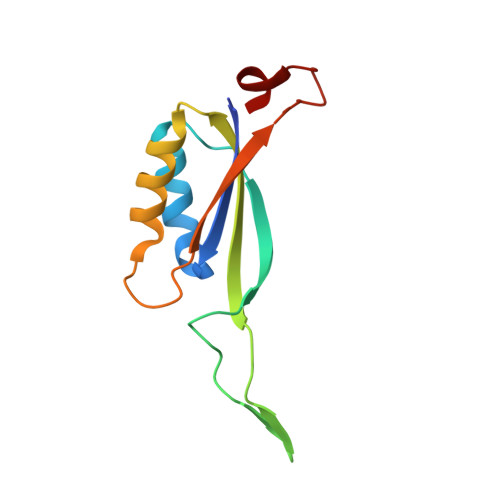
Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96
Gruswitz, F., O'Connell III, J., Stroud, R.M.(2007) Proc Natl Acad Sci U S A 104: 42-47
- PubMed: 17190799
- DOI: https://doi.org/10.1073/pnas.0609796104
- Primary Citation of Related Structures:
2NS1 - PubMed Abstract:
Ammonia conductance is highly regulated. A P(II) signal transduction protein, GlnK, is the final regulator of transmembrane ammonia conductance by the ammonia channel AmtB in Escherichia coli. The complex formed between AmtB and inhibitory GlnK at 1.96-A resolution shows that the trimeric channel is blocked directly by GlnK and how, in response to intracellular nitrogen status, the ability of GlnK to block the channel is regulated by uridylylation/deuridylylation at Y51. ATP and Mg(2+) augment the interaction of GlnK. The hydrolyzed product, adenosine 5'-diphosphate orients the surface of GlnK for AmtB blockade. 2-Oxoglutarate diminishes AmtB/GlnK association, and sites for 2-oxoglutarate are evaluated.
- Department of Biochemistry and Biophysics, Genentech Hall, School of Medicine, University of California, 600 16th Street, San Francisco, CA 94158-2517, USA.
Organizational Affiliation: