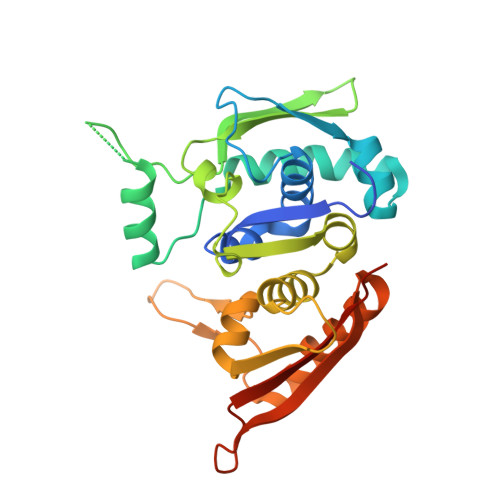
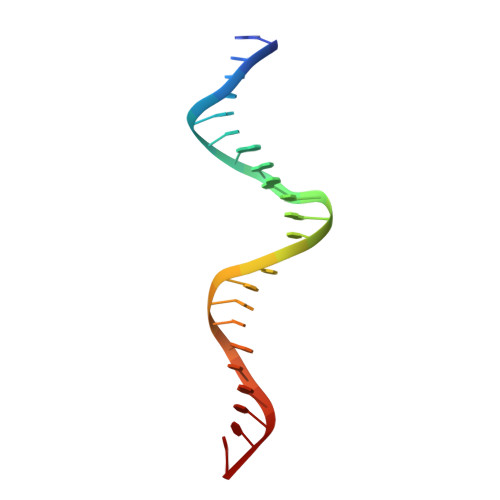
Crystal structure of pi initiator protein-iteron complex of plasmid R6K: implications for initiation of plasmid DNA replication.
Swan, M.K., Bastia, D., Davies, C.(2006) Proc Natl Acad Sci U S A 103: 18481-18486
- PubMed: 17124167
- DOI: https://doi.org/10.1073/pnas.0609046103
- Primary Citation of Related Structures:
2NRA - PubMed Abstract:
We have determined the crystal structure of a monomeric biologically active form of the pi initiator protein of plasmid R6K as a complex with a single copy of its cognate DNA-binding site (iteron) at 3.1-A resolution. The initiator belongs to the family of winged helix type of proteins. The structure reveals that the protein contacts the iteron DNA at two primary recognition helices, namely the C-terminal alpha4' and the N-terminal alpha4 helices, that recognize the 5' half and the 3' half of the 22-bp iteron, respectively. The base-amino acid contacts are all located in alpha4', whereas the alpha4 helix and its vicinity mainly contact the phosphate groups of the iteron. Mutational analyses show that the contacts of both recognition helices with DNA are necessary for iteron binding and replication initiation. Considerations of a large number of site-directed mutations reveal that two distinct regions, namely alpha2 and alpha5 and its vicinity, are required for DNA looping and initiator dimerization, respectively. Further analysis of mutant forms of pi revealed the possible domain that interacts with the DnaB helicase. Thus, the structure-function analysis presented illuminates aspects of initiation mechanism of R6K and its control.
- Department of Biochemistry and Molecular Biology, Medical University of South Carolina, Charleston, SC 29425, USA.
Organizational Affiliation: