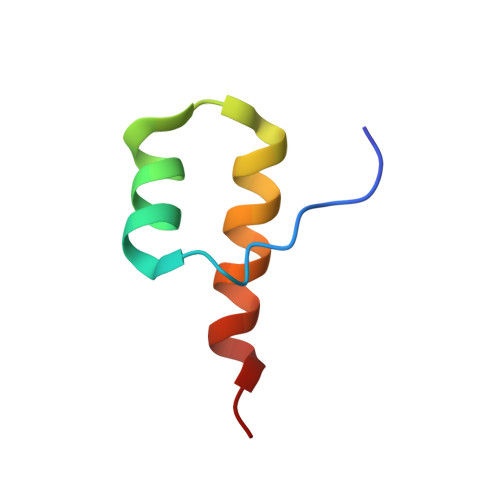
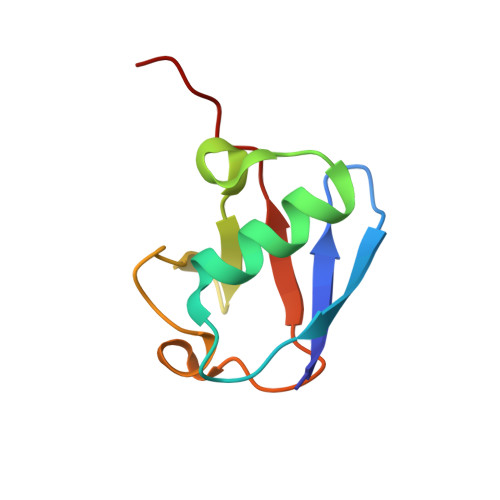
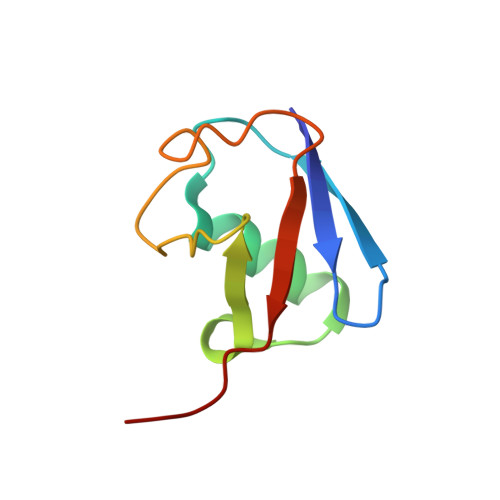
Myosin VI Contains a Compact Structural Motif that Binds to Ubiquitin Chains.
He, F., Wollscheid, H.P., Nowicka, U., Biancospino, M., Valentini, E., Ehlinger, A., Acconcia, F., Magistrati, E., Polo, S., Walters, K.J.(2016) Cell Rep 14: 2683-2694
- PubMed: 26971995
- DOI: https://doi.org/10.1016/j.celrep.2016.01.079
- Primary Citation of Related Structures:
2N0Z, 2N10, 2N11, 2N13 - PubMed Abstract:
Myosin VI is critical for cargo trafficking and sorting during early endocytosis and autophagosome maturation, and abnormalities in these processes are linked to cancers, neurodegeneration, deafness, and hypertropic cardiomyopathy. We identify a structured domain in myosin VI, myosin VI ubiquitin-binding domain (MyUb), that binds to ubiquitin chains, especially those linked via K63, K11, and K29. Herein, we solve the solution structure of MyUb and MyUb:K63-linked diubiquitin. MyUb folds as a compact helix-turn-helix-like motif and nestles between the ubiquitins of K63-linked diubiquitin, interacting with distinct surfaces of each. A nine-amino-acid extension at the C-terminal helix (Helix2) of MyUb is required for myosin VI interaction with endocytic and autophagic adaptors. Structure-guided mutations revealed that a functional MyUb is necessary for optineurin interaction. In addition, we found that an isoform-specific helix restricts MyUb binding to ubiquitin chains. This work provides fundamental insights into myosin VI interaction with ubiquitinated cargo and functional adaptors.
- Protein Processing Section, Structural Biophysics Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.
Organizational Affiliation: