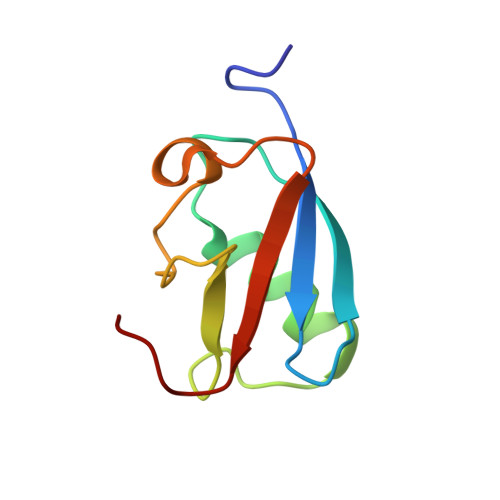
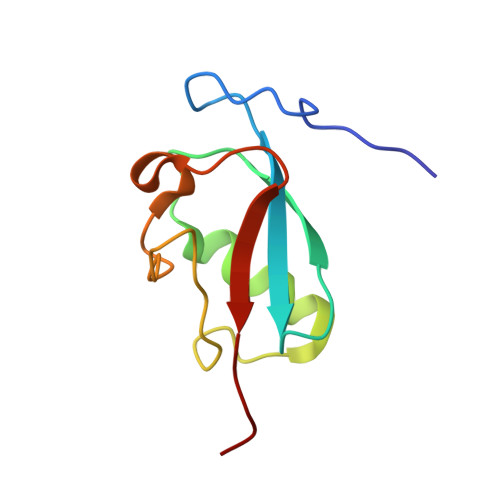
Structural insight into SUMO chain recognition and manipulation by the ubiquitin ligase RNF4.
Xu, Y., Plechanovova, A., Simpson, P., Marchant, J., Leidecker, O., Kraatz, S., Hay, R.T., Matthews, S.J.(2014) Nat Commun 5: 4217-4217
- PubMed: 24969970
- DOI: https://doi.org/10.1038/ncomms5217
- Primary Citation of Related Structures:
2MP2 - PubMed Abstract:
The small ubiquitin-like modifier (SUMO) can form polymeric chains that are important signals in cellular processes such as meiosis, genome maintenance and stress response. The SUMO-targeted ubiquitin ligase RNF4 engages with SUMO chains on linked substrates and catalyses their ubiquitination, which targets substrates for proteasomal degradation. Here we use a segmental labelling approach combined with solution nuclear magnetic resonance (NMR) spectroscopy and biochemical characterization to reveal how RNF4 manipulates the conformation of the SUMO chain, thereby facilitating optimal delivery of the distal SUMO domain for ubiquitin transfer.
- 1] Centre for Structural Biology, Department of Life Sciences, Imperial College London, South Kensington, London SW7 2AZ, UK [2].
Organizational Affiliation: