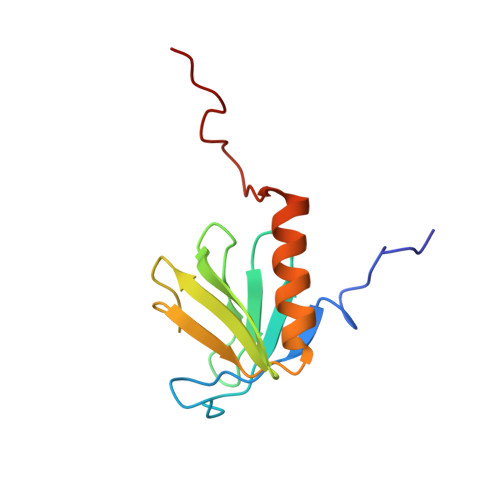
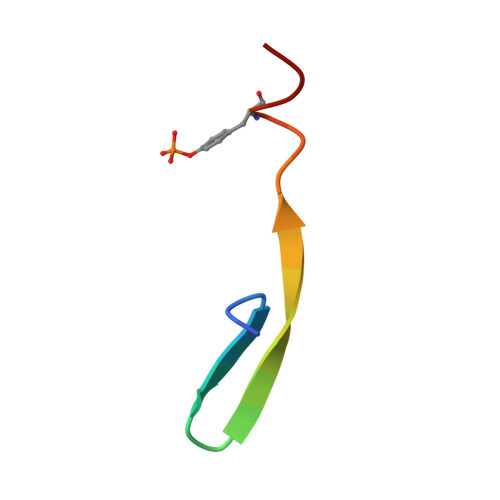
Structural insights into FRS2 alpha PTB domain recognition by neurotrophin receptor TrkB.
Zeng, L., Kuti, M., Mujtaba, S., Zhou, M.M.(2014) Proteins 82: 1534-1541
- PubMed: 24470253
- DOI: https://doi.org/10.1002/prot.24523
- Primary Citation of Related Structures:
2MFQ - PubMed Abstract:
The fibroblast growth factor receptor (FGFR) substrate 2 (FRS2) family proteins function as scaffolding adapters for receptor tyrosine kinases (RTKs). The FRS2α proteins interact with RTKs through the phosphotyrosine-binding (PTB) domain and transfer signals from the activated receptors to downstream effector proteins. Here, we report the nuclear magnetic resonance structure of the FRS2α PTB domain bound to phosphorylated TrkB. The structure reveals that the FRS2α-PTB domain is comprised of two distinct but adjacent pockets for its mutually exclusive interaction with either nonphosphorylated juxtamembrane region of the FGFR, or tyrosine phosphorylated peptides TrkA and TrkB. The new structural insights suggest rational design of selective small molecules through targeting of the two conjunct pockets in the FRS2α PTB domain.
- Department of Structural and Chemical Biology, Icahn School of Medicine at Mount Sinai, New York, New York, 10029.
Organizational Affiliation: