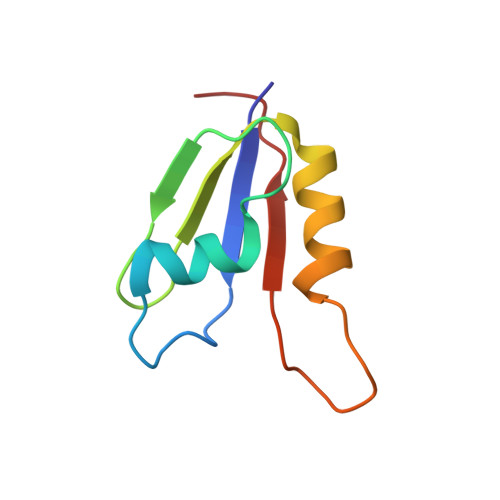
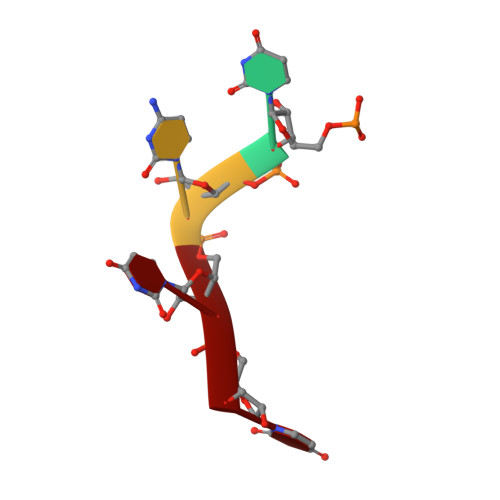
Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein
Hobor, F., Pergoli, R., Kubicek, K., Hrossova, D., Bacikova, V., Zimmermann, M., Pasulka, J., Hofr, C., Vanacova, S., Stefl, R.(2011) J Biological Chem 286: 3645-3657
- PubMed: 21084293
- DOI: https://doi.org/10.1074/jbc.M110.158774
- Primary Citation of Related Structures:
2L41 - PubMed Abstract:
Non-coding RNA polymerase II transcripts are processed by the poly(A)-independent termination pathway that requires the Nrd1 complex. The Nrd1 complex includes two RNA-binding proteins, the nuclear polyadenylated RNA-binding (Nab) 3 and the nuclear pre-mRNA down-regulation (Nrd) 1 that bind their specific termination elements. Here we report the solution structure of the RNA-recognition motif (RRM) of Nab3 in complex with a UCUU oligonucleotide, representing the Nab3 termination element. The structure shows that the first three nucleotides of UCUU are accommodated on the β-sheet surface of Nab3 RRM, but reveals a sequence-specific recognition only for the central cytidine and uridine. The specific contacts we identified are important for binding affinity in vitro as well as for yeast viability. Furthermore, we show that both RNA-binding motifs of Nab3 and Nrd1 alone bind their termination elements with a weak affinity. Interestingly, when Nab3 and Nrd1 form a heterodimer, the affinity to RNA is significantly increased due to the cooperative binding. These findings are in accordance with the model of their function in the poly(A) independent termination, in which binding to the combined and/or repetitive termination elements elicits efficient termination.
- National Centre for Biomolecular Research, Central European Institute of Technology, Faculty of Science, Masaryk University, Brno CZ-62500, Czechia.
Organizational Affiliation: